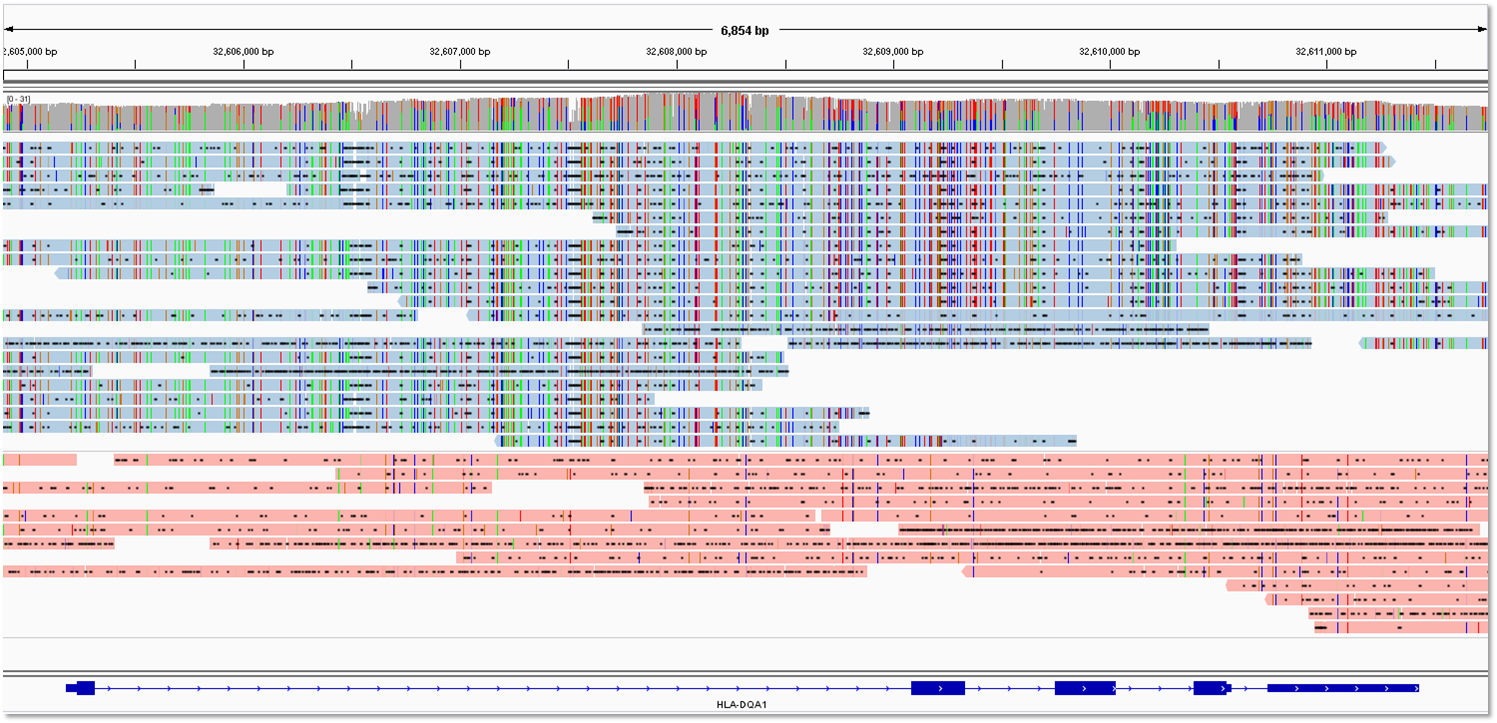
The targeted-phasing-consensus.sh script runs phasing on a subset of aligned PacBio Reads of Insert (CCS reads) corresponding to a gene or region of interest, and generates a Quiver-based consensus sequence for each phase.
This workflow was designed for experiments utilizing long fragment target capture, such as the upcoming 6 kb capture protocol using Roche NimbleGen’s® SeqCap EZ Enrichment®.
Download these two files into the same directory:
% wget https://github.com/lhon/targeted-phasing-consensus/raw/master/targeted-phasing-consensus.sh
% wget https://github.com/lhon/targeted-phasing-consensus/raw/master/faidx.zipRunning targeted-phasing-consensus.sh requires having SMRT Analysis 2.3.0 installed.
-
In SMRT Portal, use the
RS_ReadsOfInsert_Mappingprotocol with the following parameters to generate and align single best estimate Reads of Insert per molecule:- Minimum Full Passes: 0
- Minimum Predicted Accuracy: 75
-
The name of the reference,
aligned_reads.bam, andinput.fofnfrom Step 1 can then be used for the phasing step.sourcethe SMRT Analysis environment, and then runtargeted-phasing-consensus.shon a targeted region:
% source /opt/smrtanalysis/etc/setup.sh
% ./targeted-phasing-consensus.sh chr17 41243000 41244200 BRCA1 hg19_M_sorted /opt/smrtanalysis/common/jobs/087/087197/data/aligned_reads.bam /opt/smrtanalysis/common/jobs/087/087197/input.fofnIn the example above, we are looking at a subset of BRCA1 on chr17 between positions 41243000 and 41244200. hg19_M_sorted was the name of the reference in SMRT Portal, and the paths to aligned_reads.bam and input.fofn from job 87197 were specified. The results and intermediate output will be placed in the BRCA1 subdirectory.
Some relevant files in the output directory:
subset.bamis the subset of the full BAM file containing reads mapping to the region of interest.phase.outis the output from thesamtools phasecommand. Successfully phased regions are delineated by thePS(phase set) marker.M1describes detected heterogyzote SNPs that passed the filter.consensus0.fastaandconsensus1.fastaare the consensus sequences for each phase, generated by Quiver.nucmer.snpsshows the SNP differences between the phases.