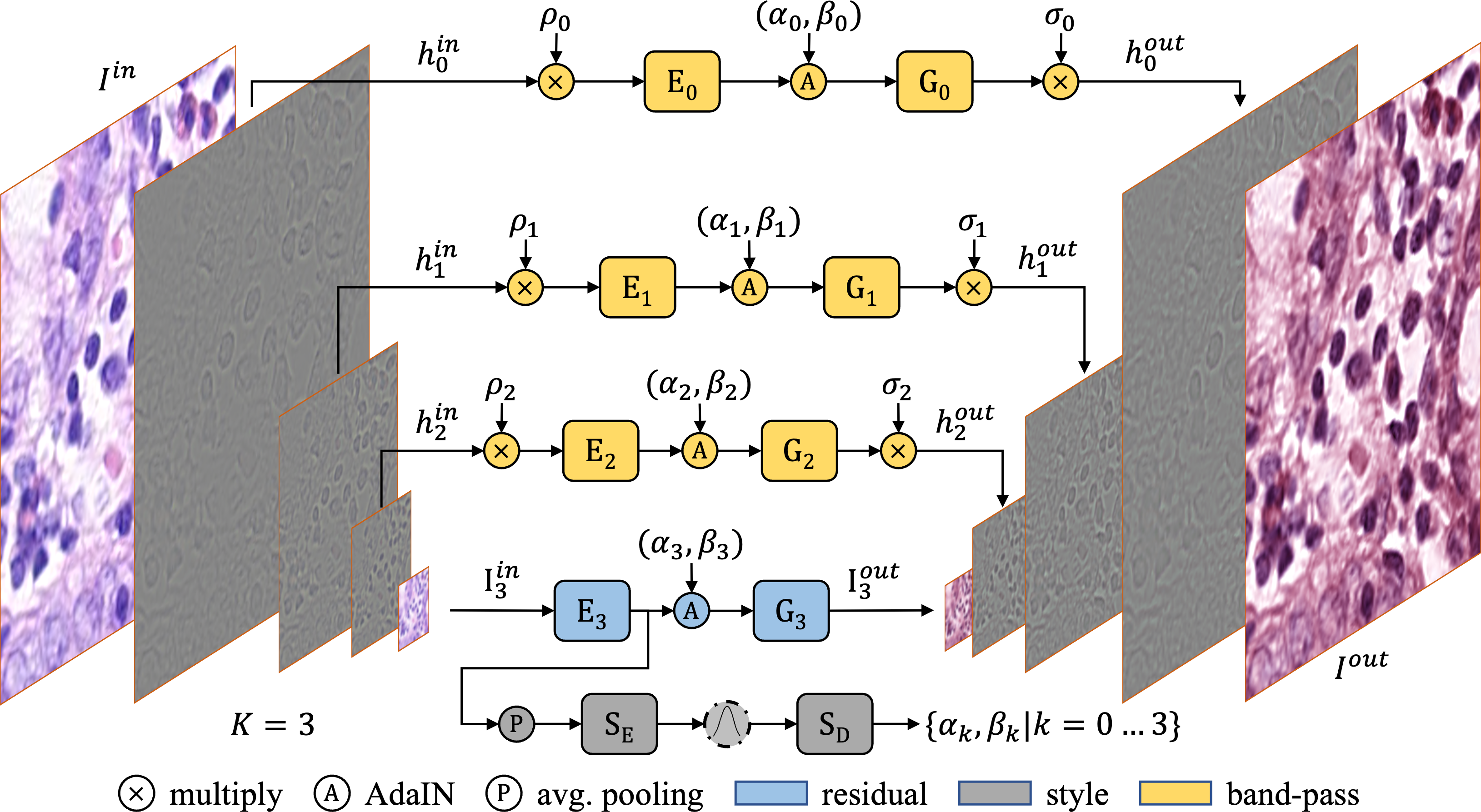
Hematoxylin and Eosin (H&E) staining is a widely used sample preparation procedure for enhancing the saturation of tissue sections and the contrast between nuclei and cytoplasm in histology images for medical diagnostics. However, various factors, such as the differences in the reagents used, result in high variability in the colors of the stains actually recorded. This variability poses a challenge in achieving generalization for machine-learning based computer-aided diagnostic tools. To desensitize the learned models to stain variations, we propose the Generative Stain Augmentation Network (G-SAN) – a GAN-based framework that augments a collection of cell images with simulated yet realistic stain variations. At its core, G-SAN uses a novel and highly computationally efficient Laplacian Pyramid (LP) based generator architecture, that is capable of disentangling stain from cell morphology. Through the task of patch classification and nucleus segmentation, we show that using G-SAN-augmented training data provides on average 15.7% improvement in F1 score and 7.3% improvement in panoptic quality, respectively. In this repo, we aim to provide a minimalistic demonstration of the G-SAN architecture and its core functionalities.
Our full paper can be found here.
Install the dependencies using:
conda create -y -n "gsan" python=3.8 ipython
conda activate gsan
conda install -y pytorch=1.7.1 torchvision cudatoolkit=10.1 -c pytorch
conda install -y scikit-image
conda install -y opencv -c conda-forge See demo.ipynb for concise examples using the pre-trained G-SAN.
This repo is built upon Imaginaire from NVIDIA.
For the full training code, please see the training branch.