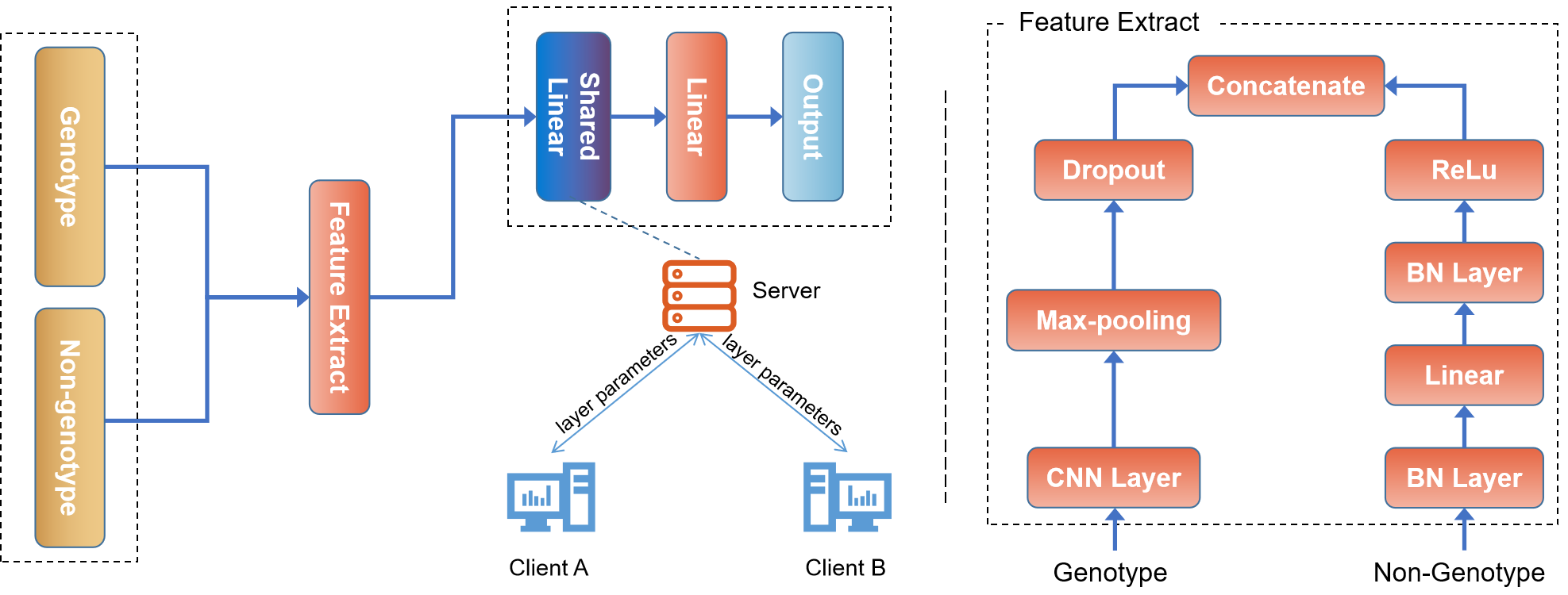
 The program 'fedgs' can be used to perform genomic selection with our proposed federated learning algorithm. In this program, phenotypes of your own data can be predicted using our algorithms. The effectiveness of FedGS has been demonstrated on a wide range of datasets, both simulated and real-world.
The program 'fedgs' can be used to perform genomic selection with our proposed federated learning algorithm. In this program, phenotypes of your own data can be predicted using our algorithms. The effectiveness of FedGS has been demonstrated on a wide range of datasets, both simulated and real-world.
Go to docker website and download the version satisfying your machine.
docker pull linjie7674/fedgs
docker run -it linjie7674/fedgs python3 dockerMain.py kfold --isFed False --genotype test Note: The test data is solely utilized for program correction testing purposes and does not hold any significance. If the aforementioned commands are executed accurately, your installation has been successful.
This project used many datasets.It contains public data and private data.
-
test data
test data is only used to test the program correction.
-
your own data
You can use your own data, which must be orginized by the follow structure.
- data - dataset_name your dataset name, which you can define it by yourself - datas.h5 this file includes your genotype data, where the first column should represent id number, and the others should be genotype data - labels_*.h5 this file includes your label data, where the first column should be the id number, and the others should be the labels corrsponding to id number. The `*` repersents that you can place different labels in your directory and you can define every labels data's name.
docker run -it -v {host directory of your own data}:/fedgs/data linjie7674/fedgs python3 dockerMain.py kfold --isFed True --genotype {local dataset name} --label_suf {local label suffix} --fed_genotyp {others dataset name} --fed_label_suf {others label suffix}Note: Supposing that user's private dataset is located in directory /home/test , the change the words above ({host directory of your own data}) to use's directory /home/test.
- isFed: train solely or federated learning
- genotype: directory name of local dataset directory
- label_suf: label suffix of local dataset
- fed_genotype: external dataset directory name, which will be - federated learning with your local data.
- fed_label_suf: label suffix of external dataset
Train without 10-fold cross validation.
docker run -it -v {host directory of your own data}:/fedgs/data linjie7674/fedgs python3 dockerMain.py train --isFed True --dataset_name {data1} --fed_dataset_name {data2} --batch_size 28 --lr 0.001 --study_name fedgsThe data used for pure training should be organized in this way.
- data
- dataset_name
- {dataset_name}_train_datas.h5
- {dataset_name}_valid_datas.h5
- {dataset_name}_test_datas.h5
Test is to get model accuracy.
docker run -it -v {host directory of your own data}:/fedgs/data linjie7674/fedgs python3 dockerMain.py test --dataset_name {directory of data} --model_path {path of trained model}Inference is to obtain trait by genotype.
docker run -it -v {host directory of your own data}:/fedgs/data linjie7674/fedgs python3 dockerMain.py infer --dataset_name {directory of data} --model_path {path of trained model}docker run -it --gpus all linjie7674/fedgs python3 dockerMain.py ...docker run -it linjie7674/fedgs python3 dockerMain.py --help