Author: Karim Makki
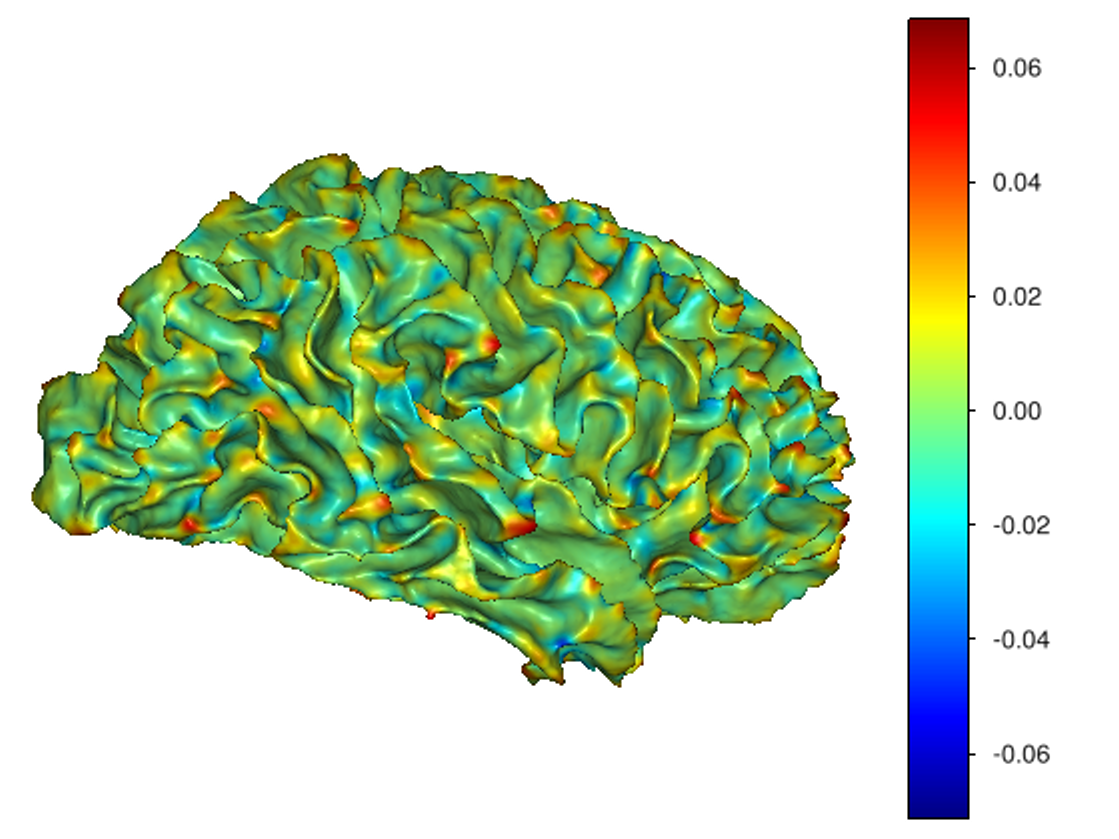
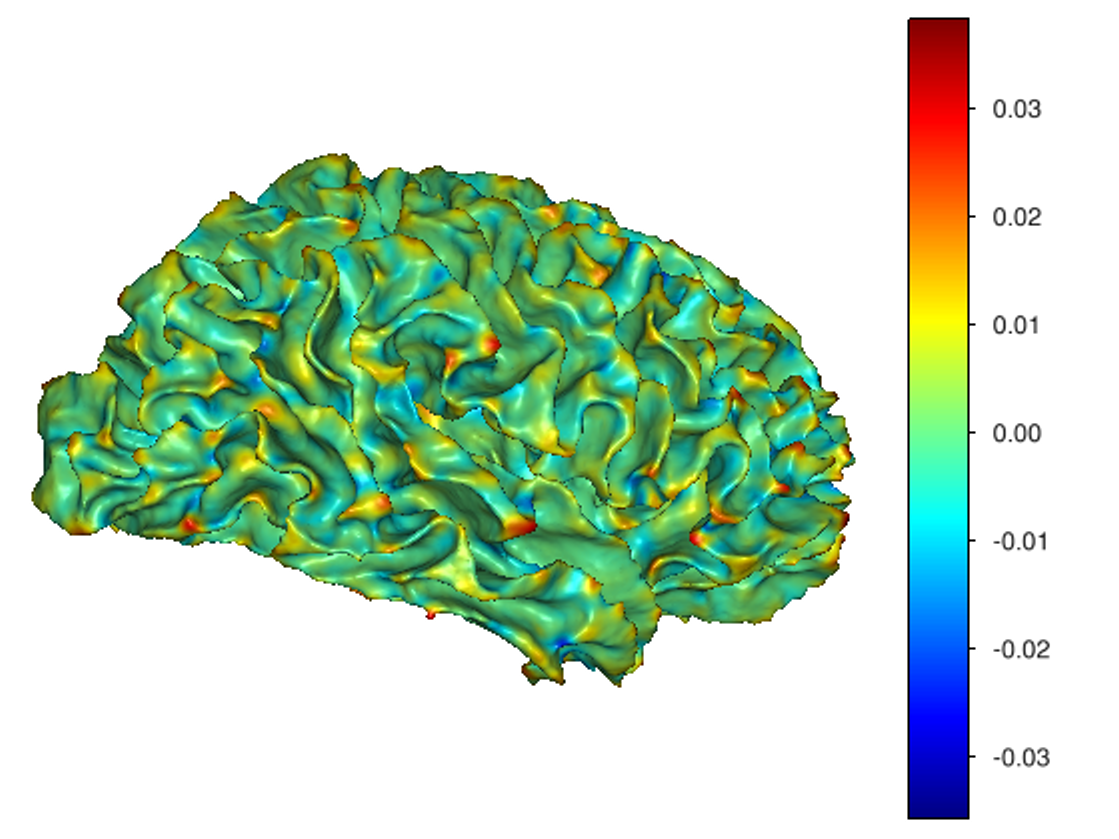
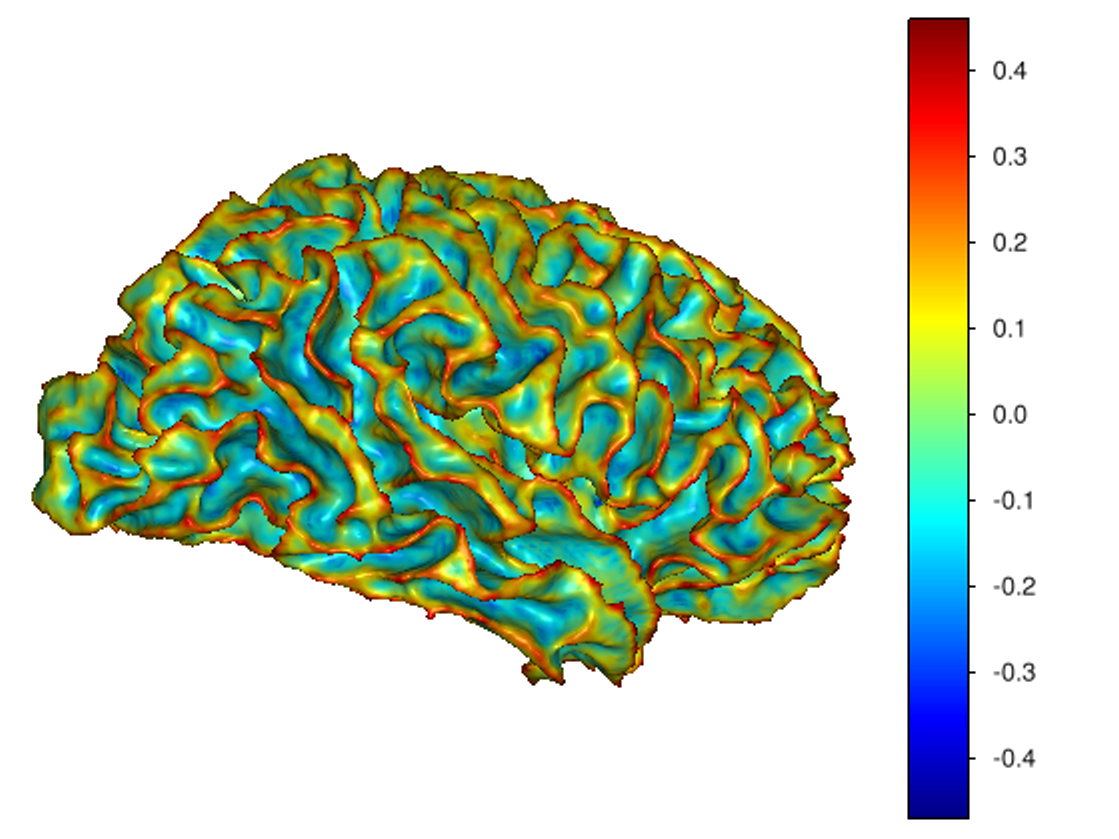
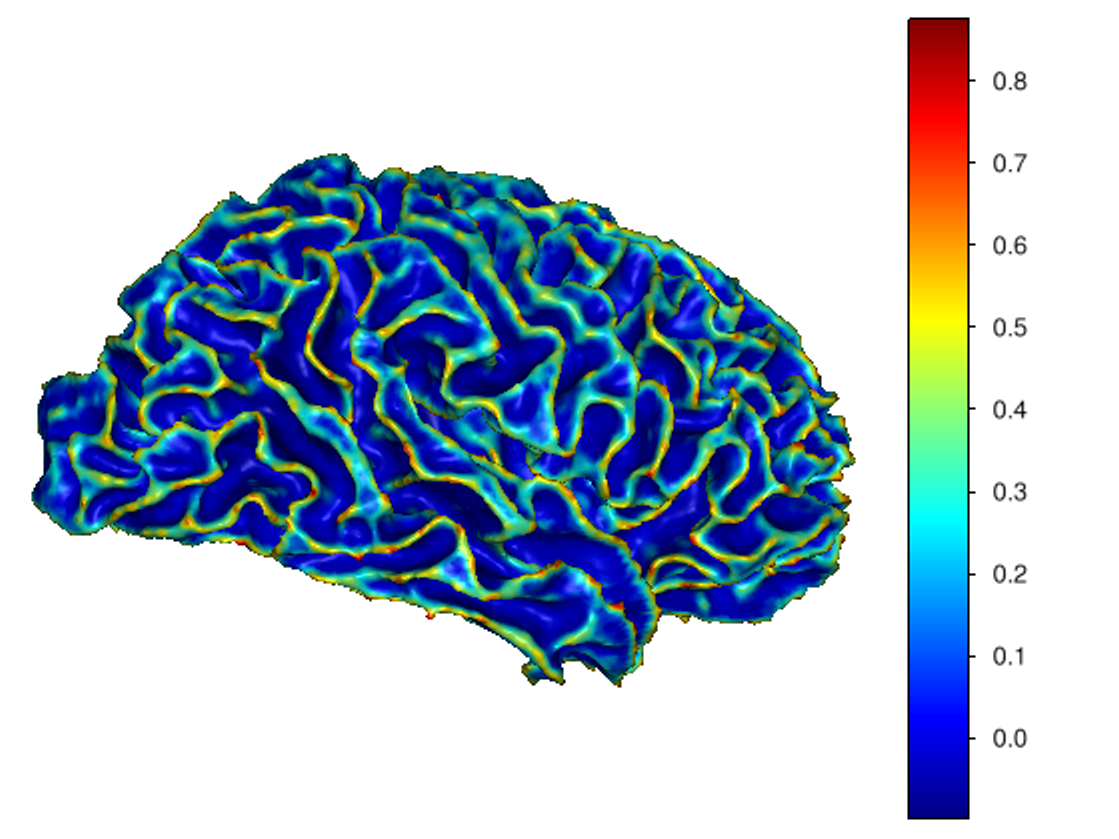
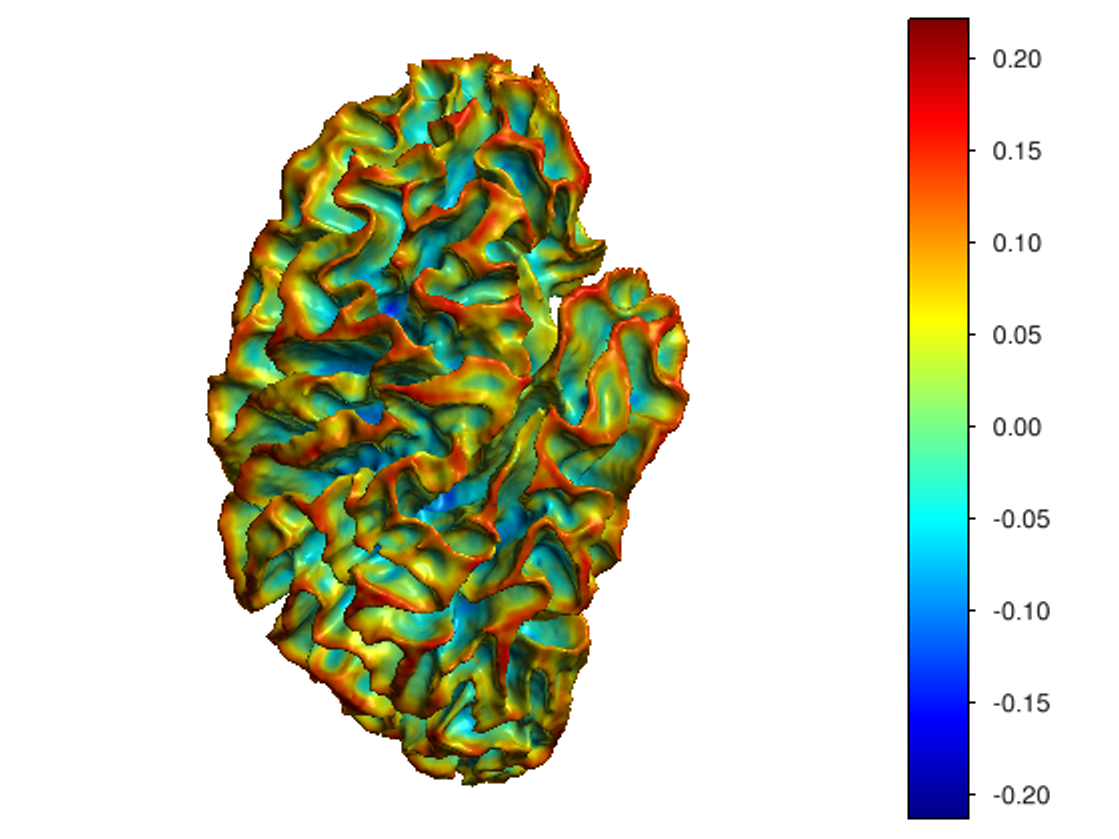
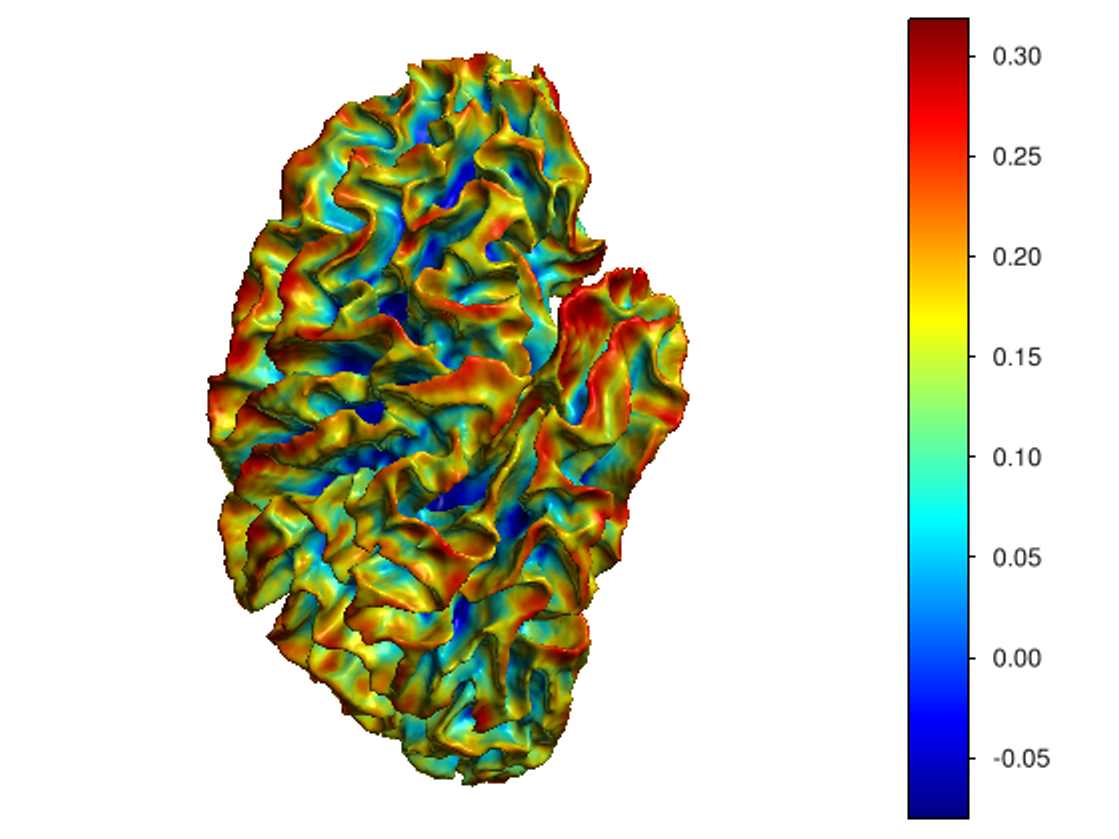
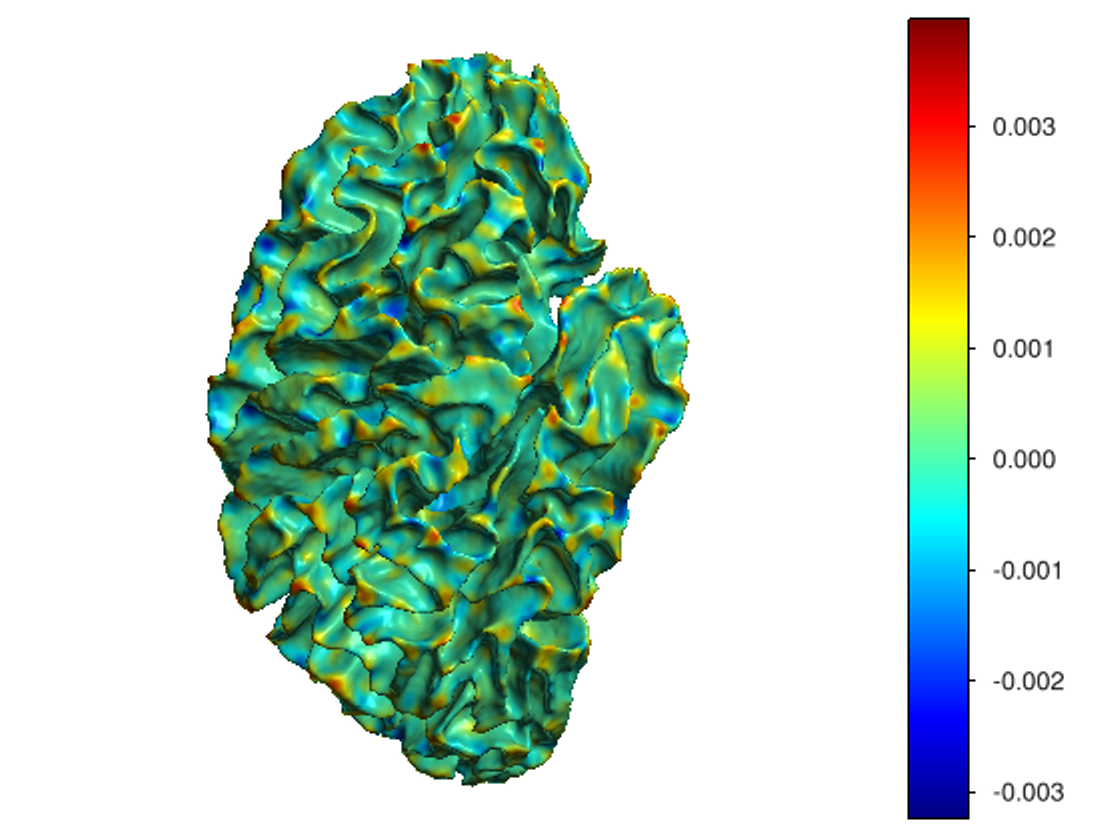
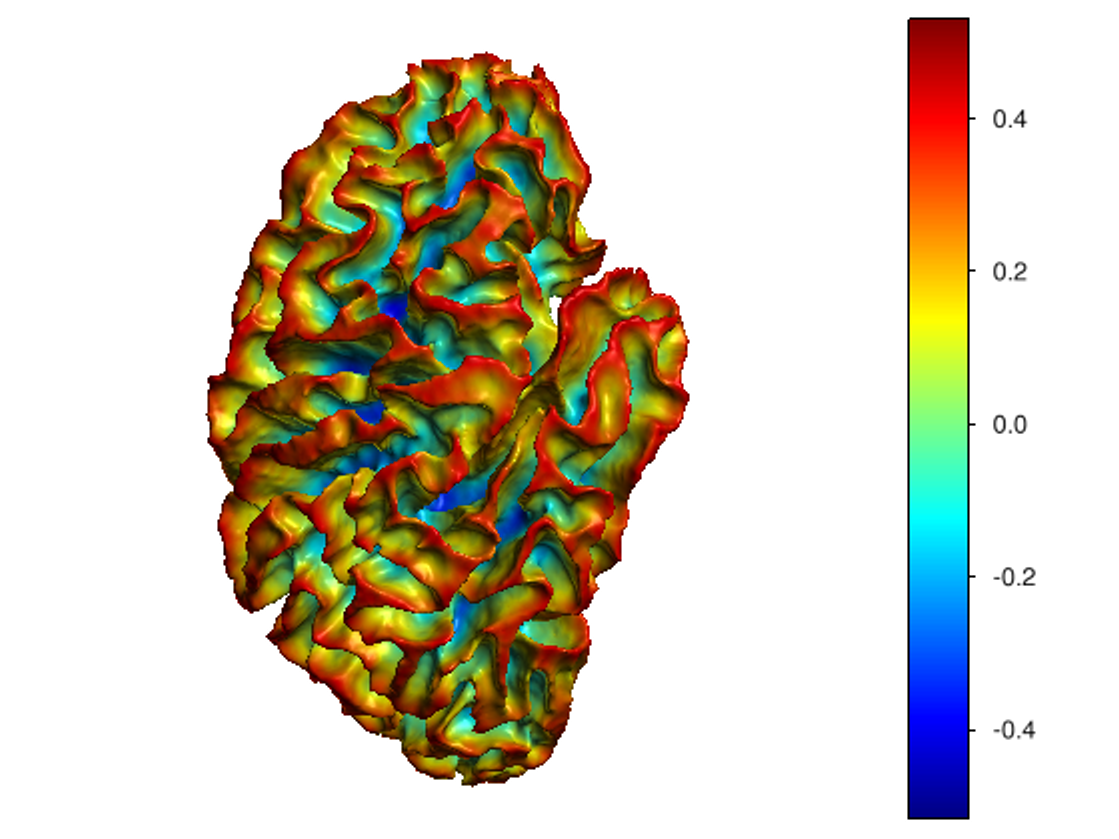
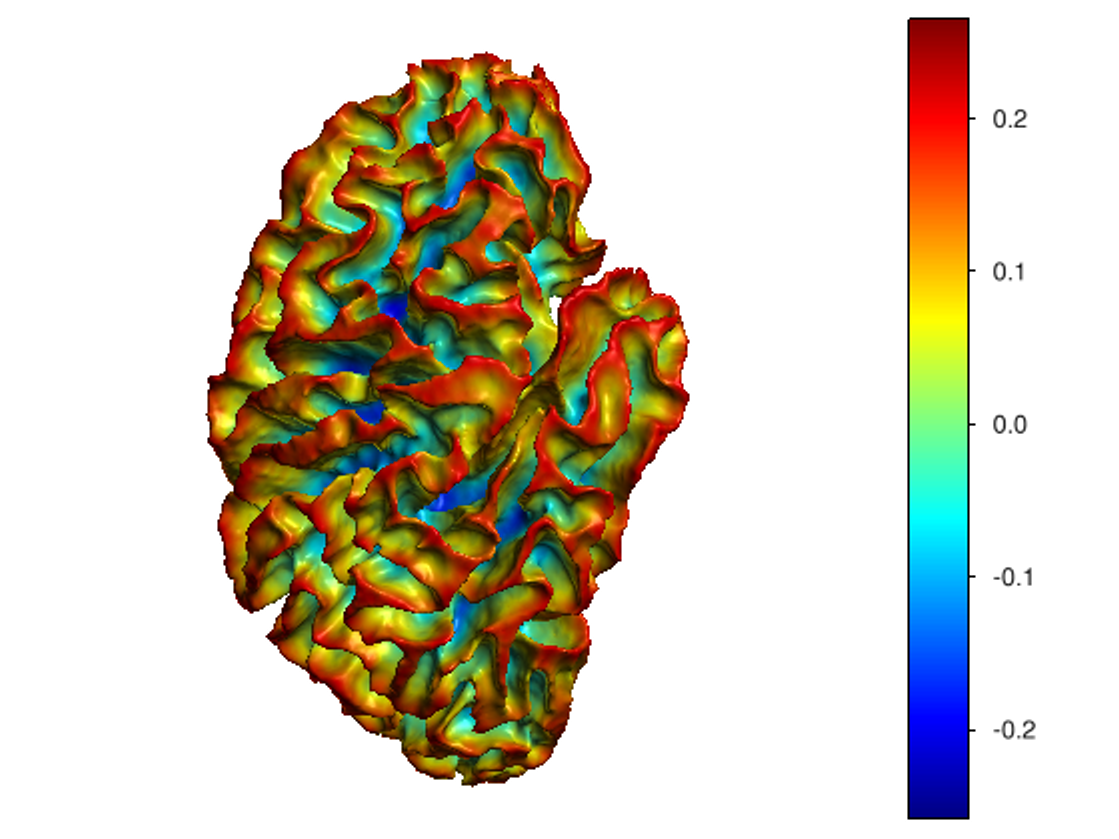
This software provides a suite of algorithms for estimating brain feature maps: curvatures (for implicit brain MRI manifolds), sulcal depth maps, and cortical thickness maps.
Inputs are essentially nifti images for binary segmentations. The user can also estimate mean and Gaussian curvature for meshes generated by Freesurfer/DHCP structural pipeline (gifti files). However, in this case, the user should also pass the corresponding nifti file as additional input to establish correspondences between real_word coordinates and vertex coordinates expressed in the image coordinate system and to perform the necessary computations in a Cartesian grid. Required softwares are available ready to run for Linux (Ubuntu) - with Windows computers being supported with a Linux Virtual Machine.
$ pip3 install visvis Trimesh nibabel pyqt5==5.14.0 rtree scikit-image
$ git clone https://github.com/k16makki/Medima_tools.git
$ cd Medima_tools
To compute all curvatures "simultaneously" (scalar, Gaussian, mean, and principal curvatures), please use one of the following commands:
curvatures_narrowband.py: to estimate curvatures directly from binary segmentations,
$ python3 curvatures_narrowband.py -in ./3D_data/white_matter.nii.gz -o ./output_folder
or,
curvatures_narrowband_4_Freesurfer_output.py: to estimate curvatures for Freesurfer output mesh,
$ python3 curvatures_narrowband_4_Freesurfer_output.py -in ./3D_data/Freesurfer_output/rh_white.nii.gz -m ./3D_data/Freesurfer_output/rh.white.gii -o ./output_folder_Freesurfer_rh
$ python3 curvatures_narrowband_4_Freesurfer_output.py -in ./3D_data/Freesurfer_output/lh_white.nii.gz -m ./3D_data/Freesurfer_output/lh.white.gii -o ./output_folder_Freesurfer_lh
To compute sulcal depth maps, please use the following commands:
$ python3 sulcal_depth.py -in ./3D_data/Freesurfer_output/rh_white.nii.gz -o ./set_your_output_directory_name
$ python3 sulcal_depth.py -in ./3D_data/Freesurfer_output/lh_white.nii.gz -o ./set_your_output_directory_name
To perform extrinsic eigen analysis, please use the following commands:
$ python3 Weingarten_map_4_Freesurfer_output.py -in ./3D_data/Freesurfer_output/rh_white.nii.gz -m ./3D_data/Freesurfer_output/rh.white.gii -o ./set_your_output_directory_name
$ python3 Weingarten_map_4_Freesurfer_output.py -in ./3D_data/Freesurfer_output/lh_white.nii.gz -m ./3D_data/Freesurfer_output/lh.white.gii -o ./set_your_output_directory_name
This software is governed by the CeCILL-B license under French law and abiding by the rules of distribution of free software. You can use, modify and/ or redistribute the software under the terms of the CeCILL-B license as circulated by CEA, CNRS and INRIA at the following URL "http://www.cecill.info".
As a counterpart to the access to the source code and rights to copy, modify and redistribute granted by the license, users are provided only with a limited warranty and the software's author, the holder of the economic rights, and the successive licensors have only limited liability.
In this respect, the user's attention is drawn to the risks associated with loading, using, modifying and/or developing or reproducing the software by the user in light of its specific status of free software, that may mean that it is complicated to manipulate, and that also therefore means that it is reserved for developers and experienced professionals having in-depth computer knowledge. Users are therefore encouraged to load and test the software's suitability as regards their requirements in conditions enabling the security of their systems and/or data to be ensured and, more generally, to use and operate it in the same conditions as regards security. The fact that you are presently reading this means that you have had knowledge of the CeCILL-B license and that you accept its terms.
If you use the code for your research, please cite the paper: "Makki. K, Ben Salem. D and Ben Amor. B. "Toward the assessment of intrinsic geometry of implicit brain MRI manifolds." IEEE Access (2021)"