A JBrowse plugin that fetches info from MyVariant.info, MyGene.info, and CIVIC and displays it.
Fetch genes and variants from hg19:
{
"label" : "MyGene.info",
"storeClass" : "MyVariantViewer/Store/SeqFeature/Genes",
"urlTemplate" : "http://mygene.info/v2/query?q=hg19.{refseq}:{start}-{end}&fields=all&email=colin.diesh@gmail.com&size=1000",
"subParts" : [
"exon"
],
"type" : "CanvasFeatures",
"hg19" : true
},
{
"label" : "MyVariant.info"
"storeClass" : "MyVariantViewer/Store/SeqFeature/Variants",
"urlTemplate" : "query?q={refseq}:{start}-{end}&size=1000&fetch_all=true&email=colin.diesh@gmail.com",
"baseUrl": "http://myvariant.info/v1/",
"type" : "CanvasFeatures",
},
{
"label" : "CIVIC",
"storeClass" : "MyVariantViewer/Store/SeqFeature/CIVIC",
"type" : "CanvasVariants",
"onClick": {
"action": "iframeDialog",
"url": "https://civicdb.org/events/genes/{gene_id}/summary/variants/{id}/summary"
}
},
Also note that a name store can be configured for querying myvariant.info via the search box as well
"names": {
"type": "MyVariantViewer/Store/Names/REST",
"url": "http://myvariant.info/v1/query"
}
Example to only query variants from clinvar (combining the exists-in clinvar query with fields=clinvar return values)
{
"storeClass" : "MyVariantViewer/Store/SeqFeature/Variants",
"baseUrl" : "http://myvariant.info/v1/",
"urlTemplate" : "query?q={refseq}:{start}-{end} AND _exists_:clinvar&size=1000&fetch_all=true&email=colin.diesh@gmail.com&fields=clinvar",
"type" : "CanvasFeatures",
"label" : "MyVariant.info clinvar"
}
See https://github.com/cmdcolin/myvariantviewer/blob/master/test/ for test data on hg19 and hg38
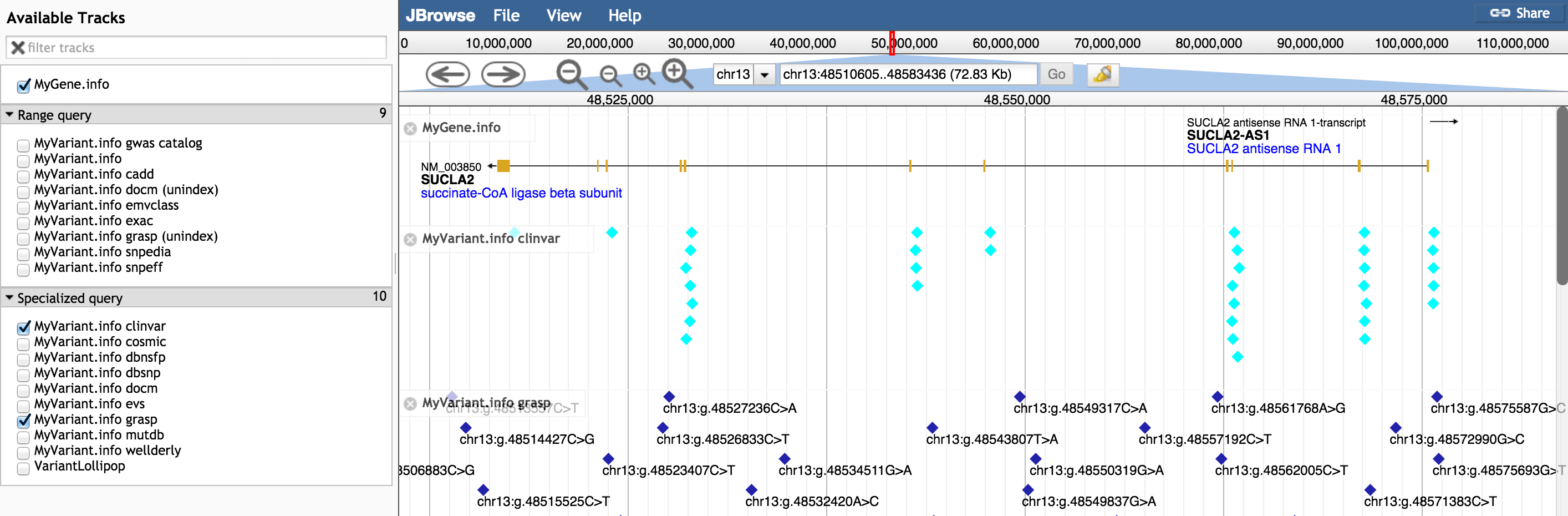
Figure. Shows the grasp and clinvar specific data tracks.
Currently tested with v2 and v3 of MyGene.info and v1 of MyVariant.info on hg19 and hg38 builds. See test/hg19 and test/hg38 for sample tracks. Also works with multiple species, see test/danio for example