Experiments with self-verification using LLMS for clinical tasks.
Code for paper "Self-Verification Improves Few-Shot Clinical Information Extraction" (Gero et al. 2023).
Extracting patient information from unstructured text is a critical task in health decision-support and clinical research. Large language models (LLMs) have shown the potential to accelerate clinical curation via few-shot in-context learning, in contrast to supervised learning, which requires costly human annotations. However, despite drastic advances, modern LLMs such as GPT-4 still struggle with issues regarding accuracy and interpretability, especially in safety-critical domains such as health. We explore a general mitigation framework using self-verification, which leverages the LLM to provide provenance for its own extraction and check its own outputs. This framework is made possible by the asymmetry between verification and generation, where the former is often much easier than the latter. Experimental results show that our method consistently improves accuracy for various LLMs across standard clinical information extraction tasks. Additionally, self-verification yields interpretations in the form of a short text span corresponding to each output, which makes it efficient for human experts to audit the results, paving the way towards trustworthy extraction of clinical information in resource-constrained scenarios.
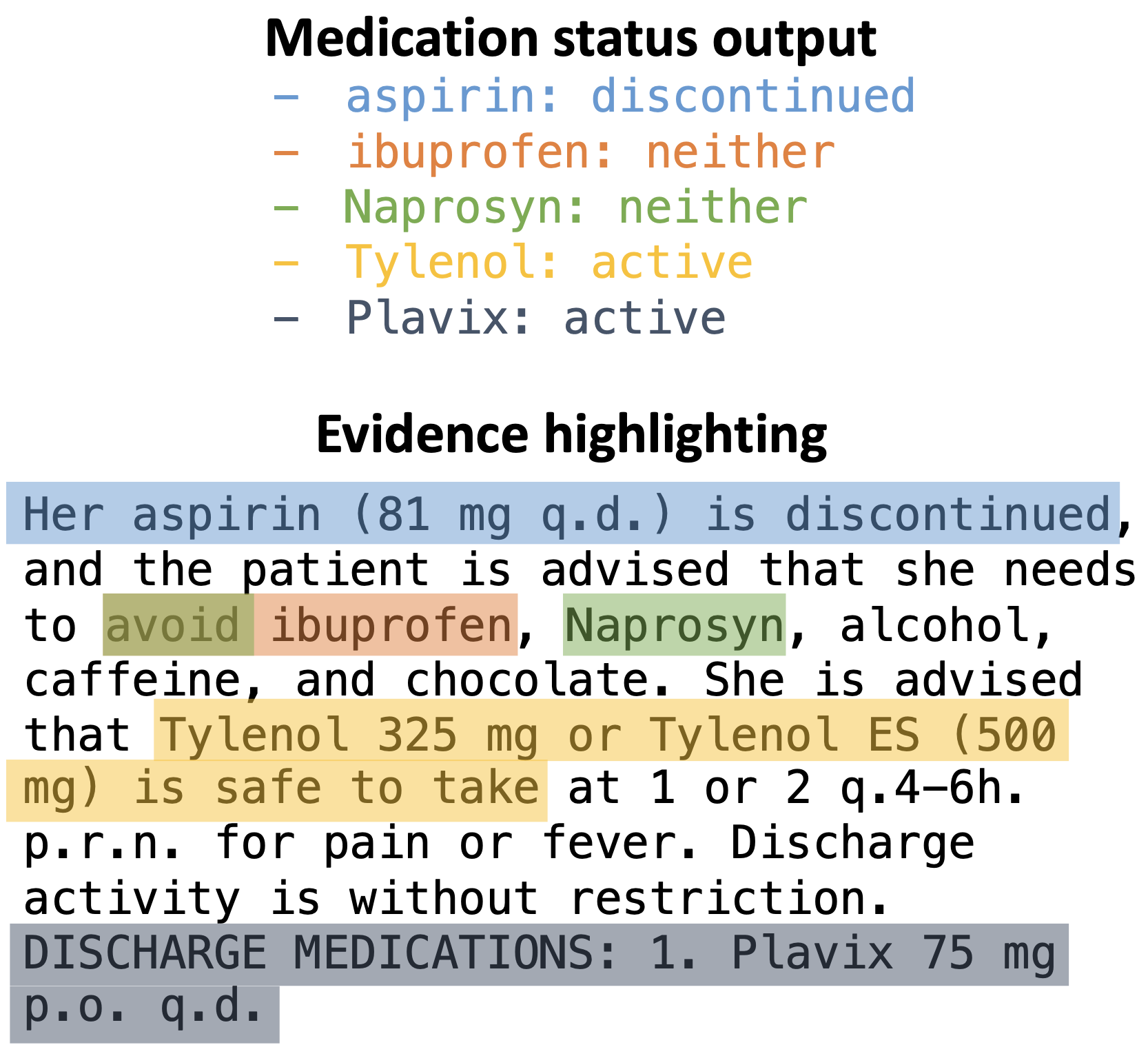
The self-verification pipeline here extracts clinical information along with evidence for each output:
@misc{gero2023selfverification,
title={Self-Verification Improves Few-Shot Clinical Information Extraction},
author={Zelalem Gero and Chandan Singh and Hao Cheng and Tristan Naumann and Michel Galley and Jianfeng Gao and Hoifung Poon},
year={2023},
journal={arXiv preprint arXiv:2306.00024},
}