Comprehensive cardiotoxicity prediction tool of small molecules on three targets: hERG, Nav1.5, Cav1.2
❗Clone first the whole repository and follow the steps bellow.
If you use CToxPred in your work, please cite the following publication:
- Issar Arab, Kristof Egghe, Kris Laukens, Ke Chen, Khaled Barakat, Wout Bittremieux, Benchmarking of Small Molecule Feature Representations for hERG, Nav1.5, and Cav1.2 Cardiotoxicity Prediction, Journal of Chemical Information and Modeling, (2023). doi:10.1021/acs.jcim.3c01301
1- Create and activate a conda environment:
$conda create -n ctoxpred python=3.7
$conda activate ctoxpred
2- Clone the repository:
$git clone git@github.com:issararab/CToxPred.git
3- Move to the repository:
$cd CToxPred
4- Install packages:
$bash install.sh
5- Run test:
$python CToxPred.py data/test_smiles_list.smi
The software saves the predictions to a CSV file named 'predictions.csv'
To re-train the models, re-evaluate the models using the same test sets, or re-run the analysis notebook, fetch first the database deposited for public use on Zenodo (https://zenodo.org/record/8245086), copy the uncompressed folder in the "data" folder, then run the notebooks.
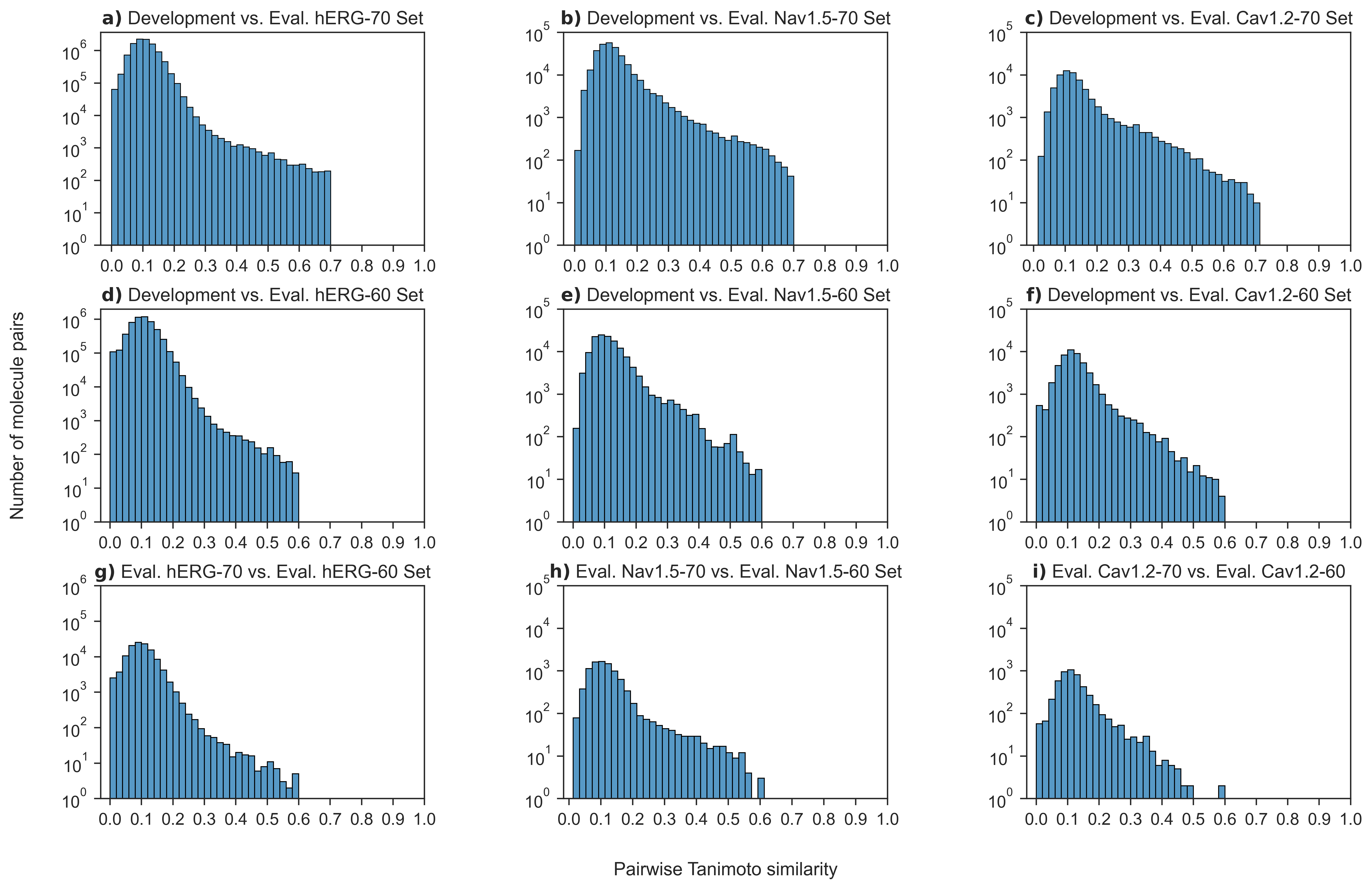
- Distribution of the pairwise Tanimoto similarity for each molecule in the datasets used for training and model evaluation
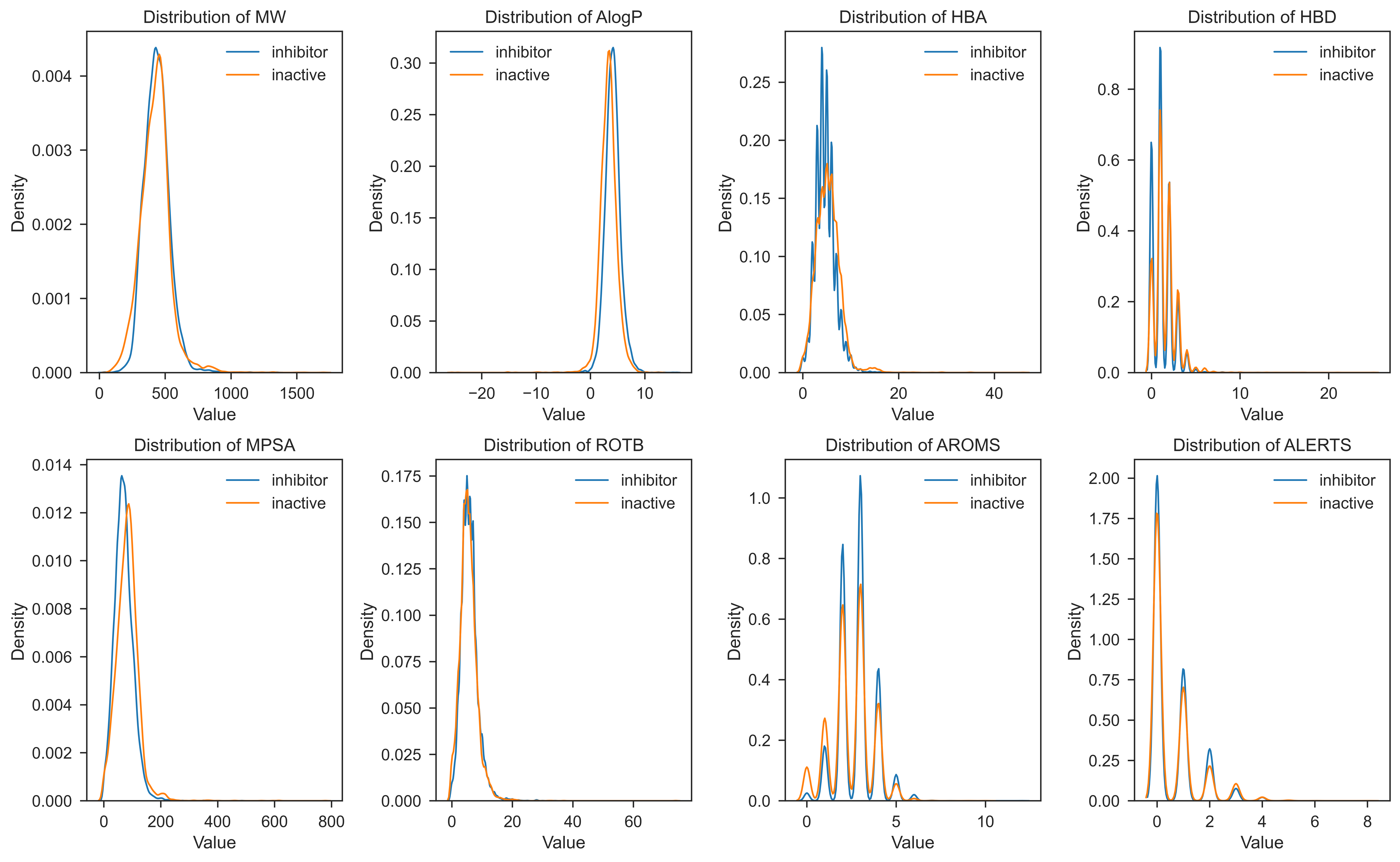
- Distributions of the 8 physicochemical properties between inhibitor(blocker) and inactive(non-blocker) compounds in the hERG dataset
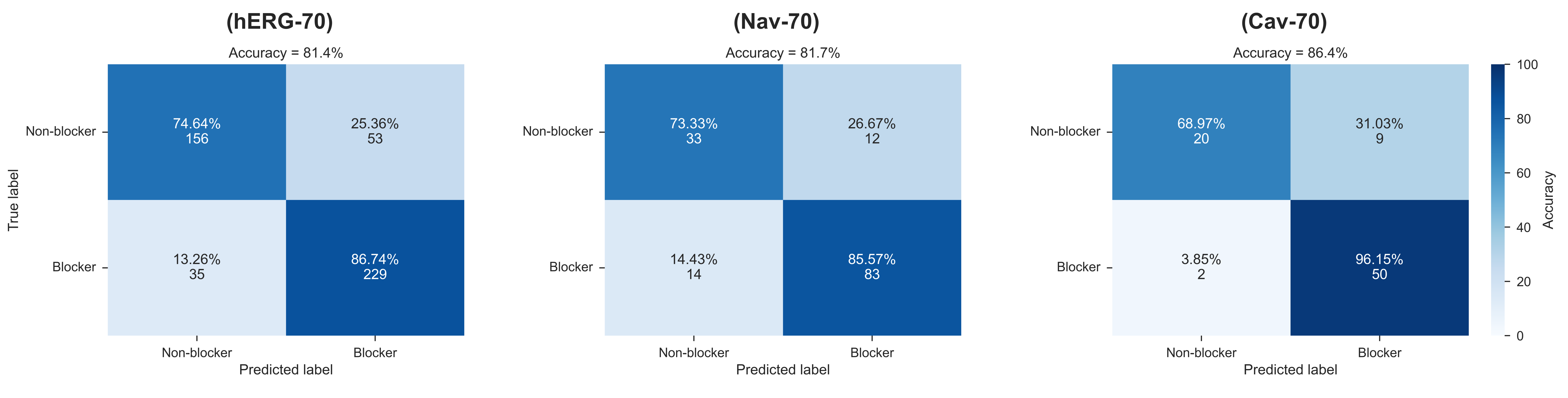
- Confusion matrix for the performance of CToxPred on all three cardiac ion channels