cannabinoid1_scra
Steps to reproduce:
- Run
align_drugs.py. This uses rdkit to generate 1000 conformers of each SCRA ligand, then aligns them to MDMB-Fubinaca from the 6N4B crystal structure. That way, the starting coordinates closely match the crystallized ligand. - Run
parameterize_drugs.py. This loads the PDB files for each ligand and parameterizes it with OpenForceField. - Run
make_complexes.sh. This runsmake_protein_drug_complex.pywhich uses parmed to blend the protein+lipid openmmSystemobject with the parameterized drugSystemobjects. Since MDMB-Fubinaca has already been removed from the protein system, the binding site is empty and can accomodate the ligand atoms without clashes. - Run
do_equilibrations.sh. This runs python scriptequilibrate.pyfor the ligands of interest. The equilibration consists of applying restraints to the protein coordinates and gradually relaxing. Relaxation of restraints means every 12psrestraint = 0.9*restraint. Typically this workflow also applies restraints to the drugs, but we found, due to differences in shape between crystallized MDMB-Fubinaca and the drugs used here, that the restraints held the ligands a little too close to the protein atoms and led to occasional NaNs. - Run
do_productions.sh. This runs python scriptproduction.pywhich simulates for 90ns using OpenMM.
Results:
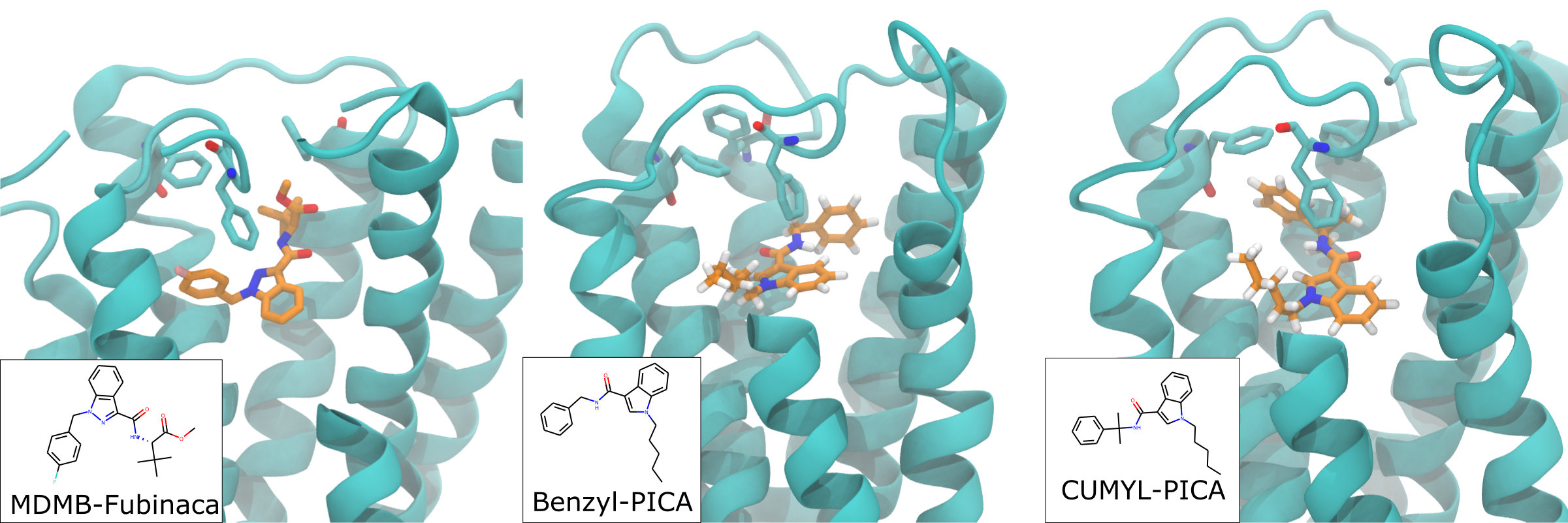
We simulated four ligands with pentyl tails to determine the mechanism of the observed SAR. In short, the crystallized ligand conformation is either maintained or lost depending on the level of steric freedom in the amino group that links the phenyl 'headgroup' and the indole scaffold 'body'. The in vitro data show that the CUMYL linker, i.e. the one with two methyls and therefore the most restricted, has the highest affinity to CB1. Conversely, the benzyl linker, i.e. the one with no methyls and the least restricted, has the lowest affinity. The change in steric freedom does not simply lead to a change in RMSD that explains binding affinity, but instead leads to a change in conformation away from that observed in the crystal structure. See below for representative snapshots of these two ligands in the CB1 receptor. The CUMYL ligand closely matches the crystallized conformation, while the benzyl ligand has rotated the headgroup around the amino linker to pull the phenyl headgroup onto the opposite side of the binding vestibule. Both are stable in these conformations with approximately equal RMSD.
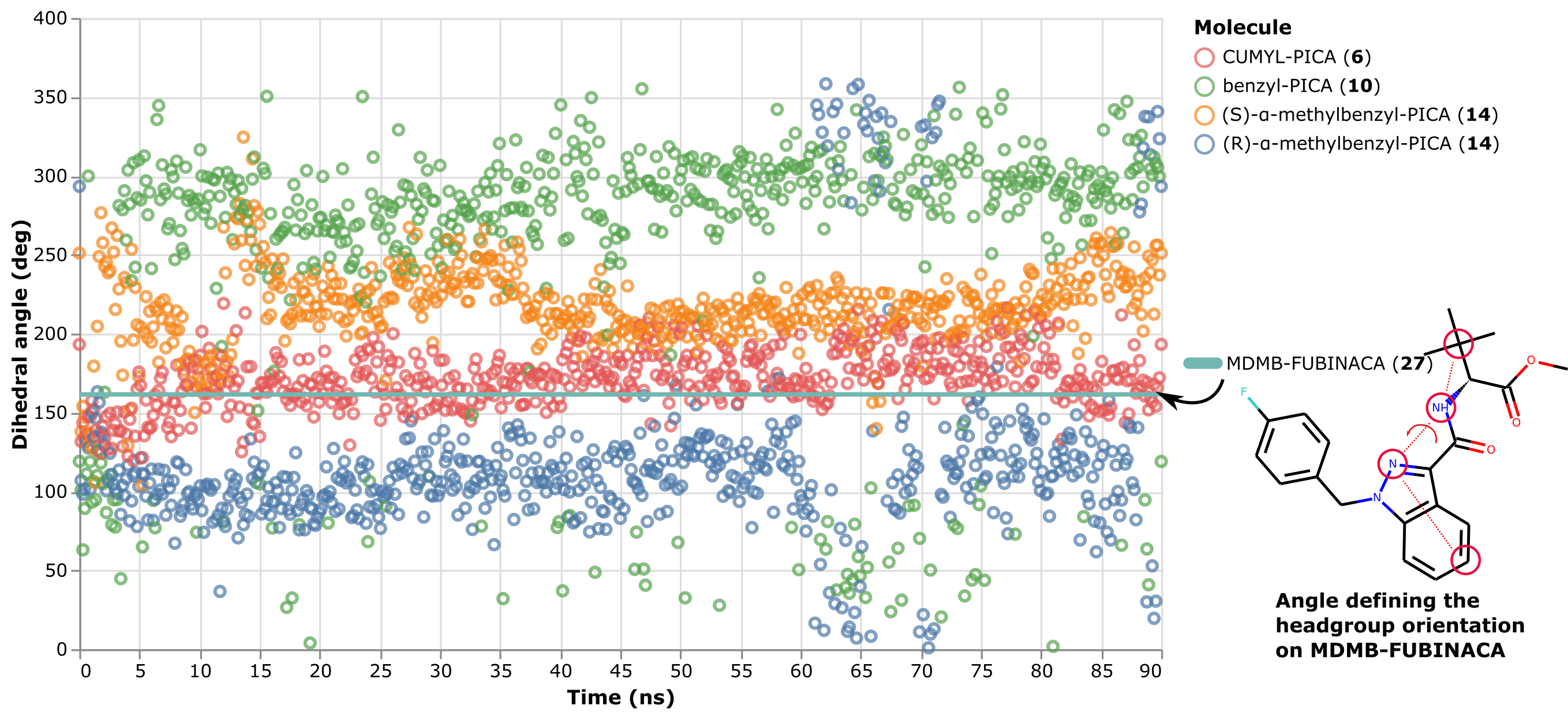
The two other ligands have (R)- or (S)- chiral single methyls. The in vitro data shows these have intermediate affinity compared with the CUMYL and benzyl linkers. The simulation data show that the headgroup has intermediate orientations compared to the CUMYL and benzyl ligands, leading to a model where the level of steric freedom in the linker group dictates the position of the headgroup region, with the most restricted ligand perfectly recapitulating the crystal structure conformation (shown as a horizontal bar in the graph below) and the other ligands having close-to or far-away conformations to the same degree as their binding affinity. See below for time traces of the orientation of the headgroup to see this pattern. The orientation is measured by a torsional angle defined across the indole scaffold and headgroup region: