An HP 2D Lattice Environment with a Gym-like API for the protein folding problem
This is a Python library that formulates Lau and Dill's (1989) hydrophobic-polar two-dimensional lattice model as a reinforcement learning problem. It follows OpenAI Gym's API, easing integration for reinforcement learning solutions. This library only implements a two-dimensional square lattice, but different lattice structures will be done in the future.
Screenshots from render():
- numpy==1.14.2
- gym==0.10.4
- six==1.11.0
- pytest==3.2.1 (dev)
- setuptools==39.0.1 (dev)
This package is only compatible with Python 3.4 and above. To install this package, please follow the instructions below:
- Install OpenAI Gym and its dependencies.
- Install the package itself:
git clone https://github.com/ljvmiranda921/gym-lattice.git
cd gym-lattice
pip install -e .
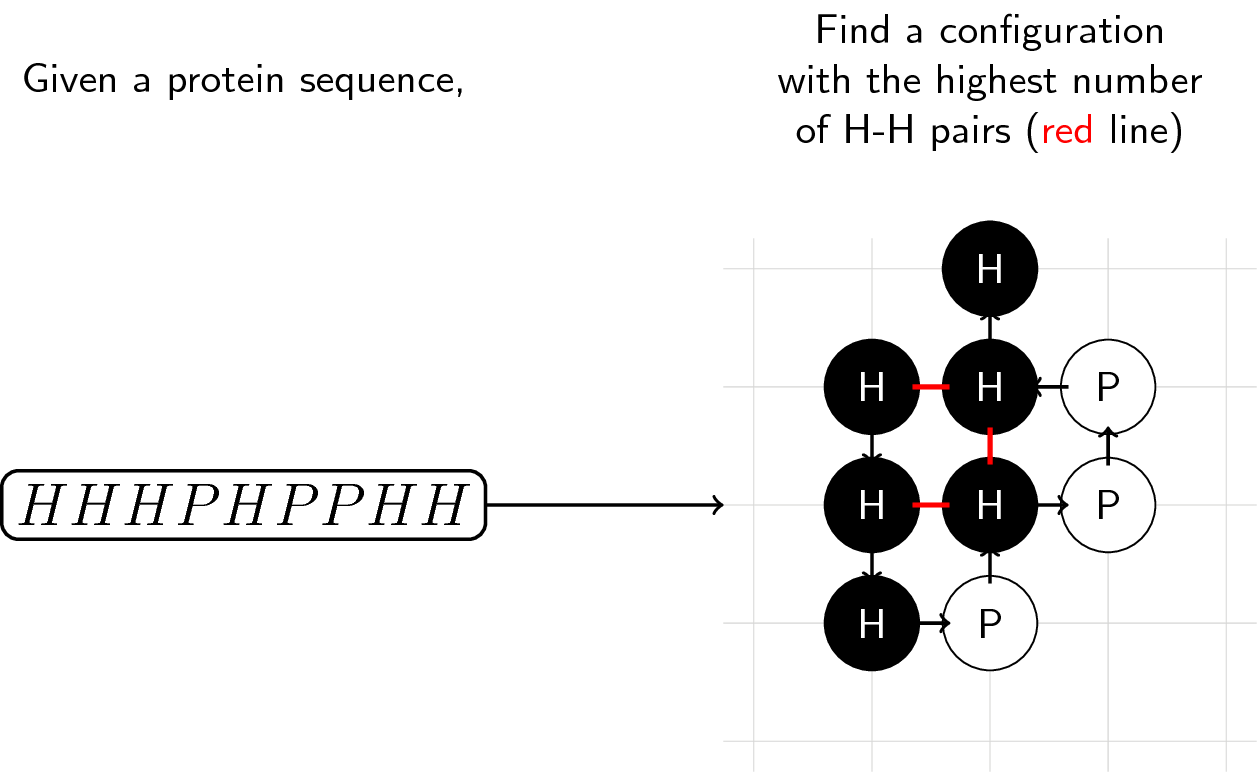
Given a sequence of H and P molecules, find a configuration with the highest number of adjacent H-H pairs. Your base score is determined by the number of H-H pairs you can create.
As with most "games", there are some rules that must be observed when folding proteins. We then pattern our rules from Dill and Lau's (1989) lattice statistical mechanics for protein conformation. For our purposes, here it is in its simplest form:
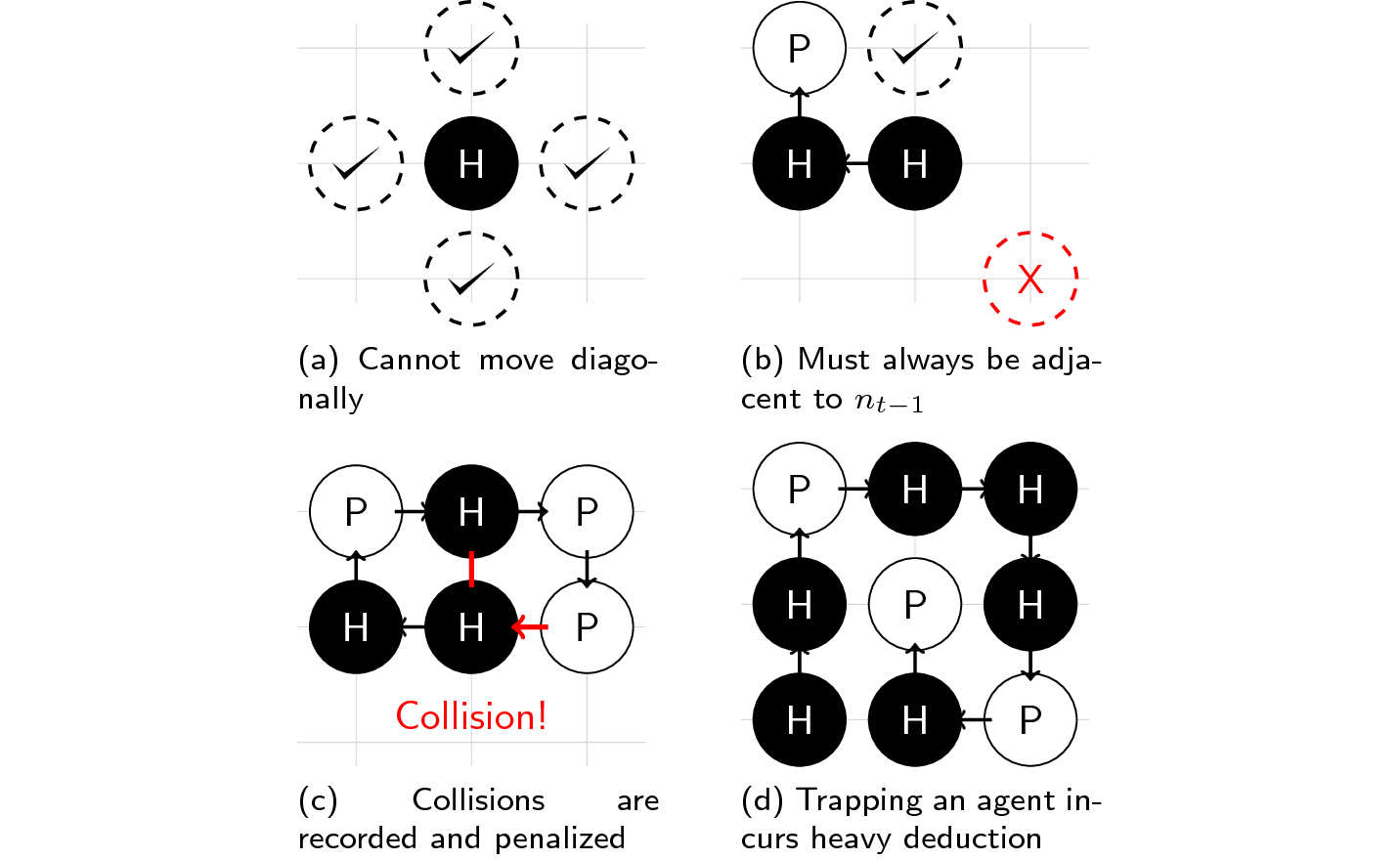
- You can only perform the following actions: left, down, up, or right.
- You can only put a molecule adjacent to the previous one you've placed.
- Assigning a molecule to an occupied space will incur a penalty.
- Trapping yourself and running out of moves will result in a heavy deduction.
Initializing the environment requires the protein sequence of type str.
In addition, you can also set the amount of penalty incurred during a
collision or whenever the agent is trapped.
Let's try this out with a random agent on the protein sequence HHPHH!
from gym_lattice.envs import Lattice2DEnv
from gym import spaces
import numpy as np
np.random.seed(42)
seq = 'HHPHH' # Our input sequence
action_space = spaces.Discrete(4) # Choose among [0, 1, 2 ,3]
env = Lattice2DEnv(seq)
for i_episodes in range(5):
env.reset()
while not env.done:
# Random agent samples from action space
action = action_space.sample()
obs, reward, done, info = env.step(action)
env.render()
if done:
print("Episode finished! Reward: {} | Collisions: {} | Actions: {}".format(reward, info['collisions'], info['actions']))
breakSample output:
Episode finished! Reward: -2 | Collisions: 1 | Actions: ['U', 'L', 'U', 'U']
Episode finished! Reward: -2 | Collisions: 0 | Actions: ['D', 'L', 'L', 'U']
Episode finished! Reward: -2 | Collisions: 0 | Actions: ['R', 'U', 'U', 'U']
Episode finished! Reward: -3 | Collisions: 1 | Actions: ['U', 'L', 'D', 'D']
Episode finished! Reward: -2 | Collisions: 2 | Actions: ['D', 'R', 'R', 'D']
Your agent can perform four possible actions: 0 (left), 1 (down), 2
(up), and 3 (right). The number choices may seem funky at first but just
remember that it maps to the standard vim keybindings.
Observations are represented as a 2-dimensional array with 1 representing
hydrophobic molecules (H), -1 for polar molecules (P), and 0 for free
spaces. If you wish to obtain the chain itself, you can do so by accessing
info['state_chain'].
An episode ends when all polymers are added to the lattice OR if the sequence of actions traps the polymer chain (no more valid moves because surrounding space is fully-occupied). Whenever a collision is detected, the agent should enter another action.
This environment computes the reward in the following manner:
# Reward at timestep t
reward_t = state_reward + collision_penalty + trap_penalty- The
state_rewardis the number of adjacent H-H molecules in the final state. In protein folding, the state_reward is synonymous to computing the Gibbs free energy, i.e., thermodynamic assumption of a stable molecule. Its value is 0 in all timesteps and is only computed at the end of the episode. - The
collision_penaltyat timestep t accounts for collision events whenever the agent chooses to put a molecule at an already-occupied space. Its default value is -2, but this can be adjusted by setting thecollision_penaltyat initialization. - The
trap_penaltyis only computed whenever the agent has no more moves left and is unable to finish the task. The episode ends, thus computing thestate_reward, but subtracts a deduction dependent on the length of the actual sequence.
Are you using gym-lattice in your paper or project? Cite us!
@misc{miranda2018gymlattice,
author = {Lester James V. Miranda},
title = {gym-lattice: an HP 2D Lattice Environment with a
Gym-like API for the protein folding problem},
month = apr,
year = 2018,
doi = {10.5281/zenodo.1214967},
url = {https://doi.org/10.5281/zenodo.1214967}
}