Detecting events in sleeping tinnitus patients
Work for SIOPI, by Robin Guillard & Louis Korczowski (2020-)
This toolbox aims at detecting specific physiological events during sleep of patient suffering of tinnitus. For now, the main focus is
- EMG: automatic bruxism evaluation using detection of burst in electromyography activity
- MEMA: automatic middle-ear muscles activation using detection of pressure variation of the ear canal
To so so, this toolbox is organized in several modules and folders:
TINNSLEEP
- config: configuration for cross-module usage storing user-specific files, folders and data. It is REQUIRED to add your data folders (hardcoded) if you want to use the preconfigured script and notebooks.
- data: load, prepare and annotate data (mainly .edf using
mne) - utils: a lot of useful methods for preparing data, labels by doing simple operations
- signal: signal processing and automatic artifact thresholding
- classification: classification methods to detects events and artifacts. The main method is AMDT (Adaptive Mean Amplitude Thresholding).
- pipeline: "ready-to-go" configured ADMT pipeline for event classification
- events: methods to build and differentiate events
- reports: automatic reporting system using all above
SCRIPT
compute_results.pyis a preconfigured script which should be used in command line with option (by default it WON'T overwrite exiting results). It will load all suitable data and usedata_info.csvto infers which operation to do for which file. Requirements: (a) add your local data folders intinnsleep.config.Config(b) have a configureddata_info.csvfor the each related file.
NOTEBOOKS
- Bruxism_detection: preconfigured notebook to classify and visualize classification outputs for Bruxism detection of one subject.
- Bruxism_Inter_subject_analyze: preconfigured notebook for group-level analyze of bruxism (results are computed using
compute_results.py) - Middle_ear_detection: preconfigured notebook to classify and visualize classification outputs for middle ear muscles activity (MEMA) detection
- Middle_ear_Inter_subject_analyze: preconfigured notebook for group-level analyze of MEMA (results are computed using
compute_results.py) - and more...
As shown in notebooks/demo_mema_detection.ipynb (e.g. Figure 1), a standardized method is used for MEMA and EMG events detection using the tinnsleep.pipeline methods. Here is some explained results.
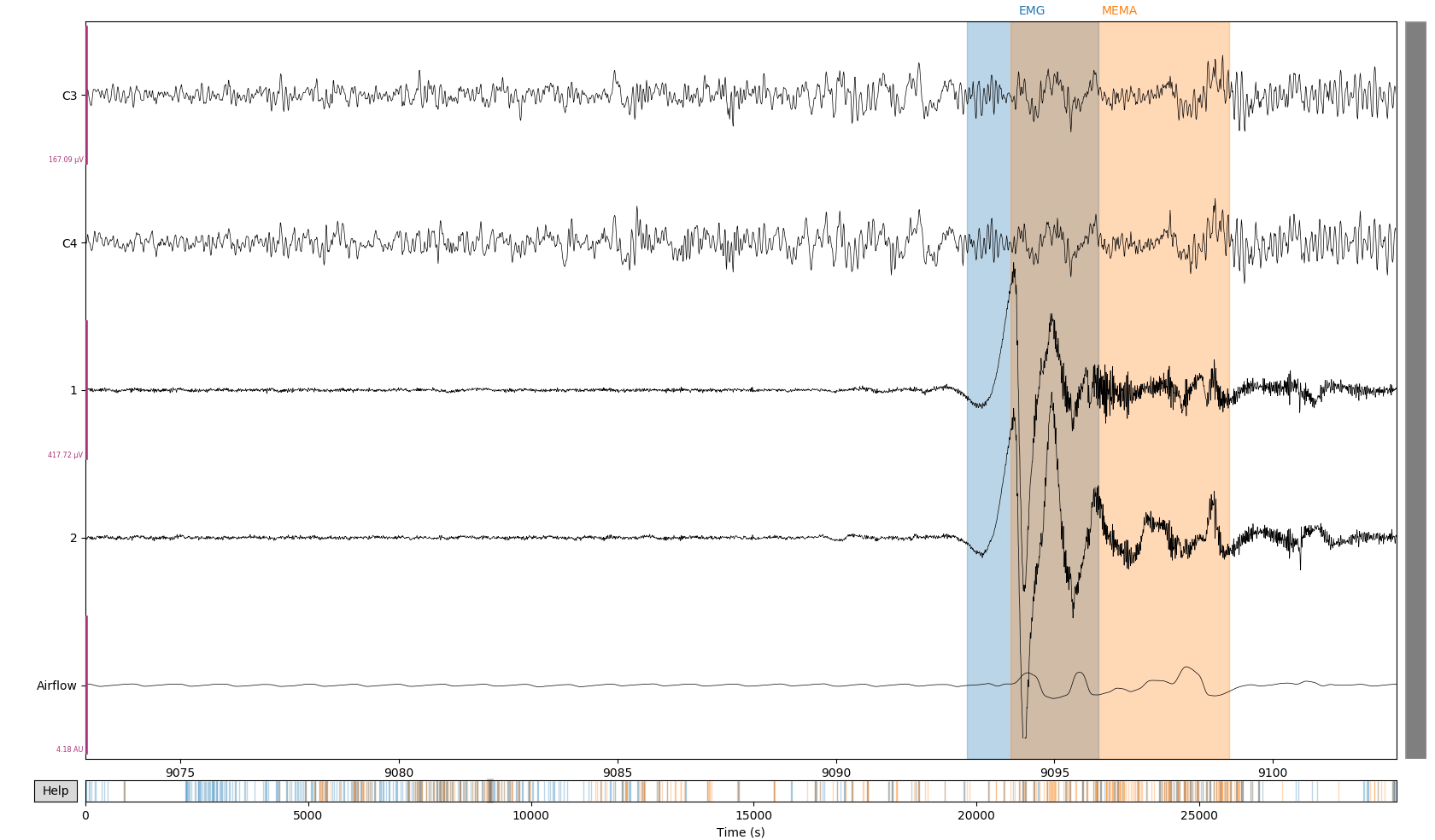 Figure 1: Detection of both MEMA (orange) and EM (blue) events using adaptive scheme. In this situation, EMG baseline has abruptly changed but the adaptive thresholding is able to both detect the first burst of adapt to the new baseline (right). Meanwhile a burst of several MEMA is detected.
Figure 1: Detection of both MEMA (orange) and EM (blue) events using adaptive scheme. In this situation, EMG baseline has abruptly changed but the adaptive thresholding is able to both detect the first burst of adapt to the new baseline (right). Meanwhile a burst of several MEMA is detected.
Classification is not enough to categorize events (see Figure 2), the tinnsleep.events methods allow to differentiate between different types of events. To do so, events are labeled into bursts and episodes:
burstsare continuous events which are classified using thetinnsleep.classificationmodule.episodesare a succession of bursts which are merged together using scoring methods. Eachepisodeis labeled thanks to the properties of his bursts into phasic, tonic or mixed events.
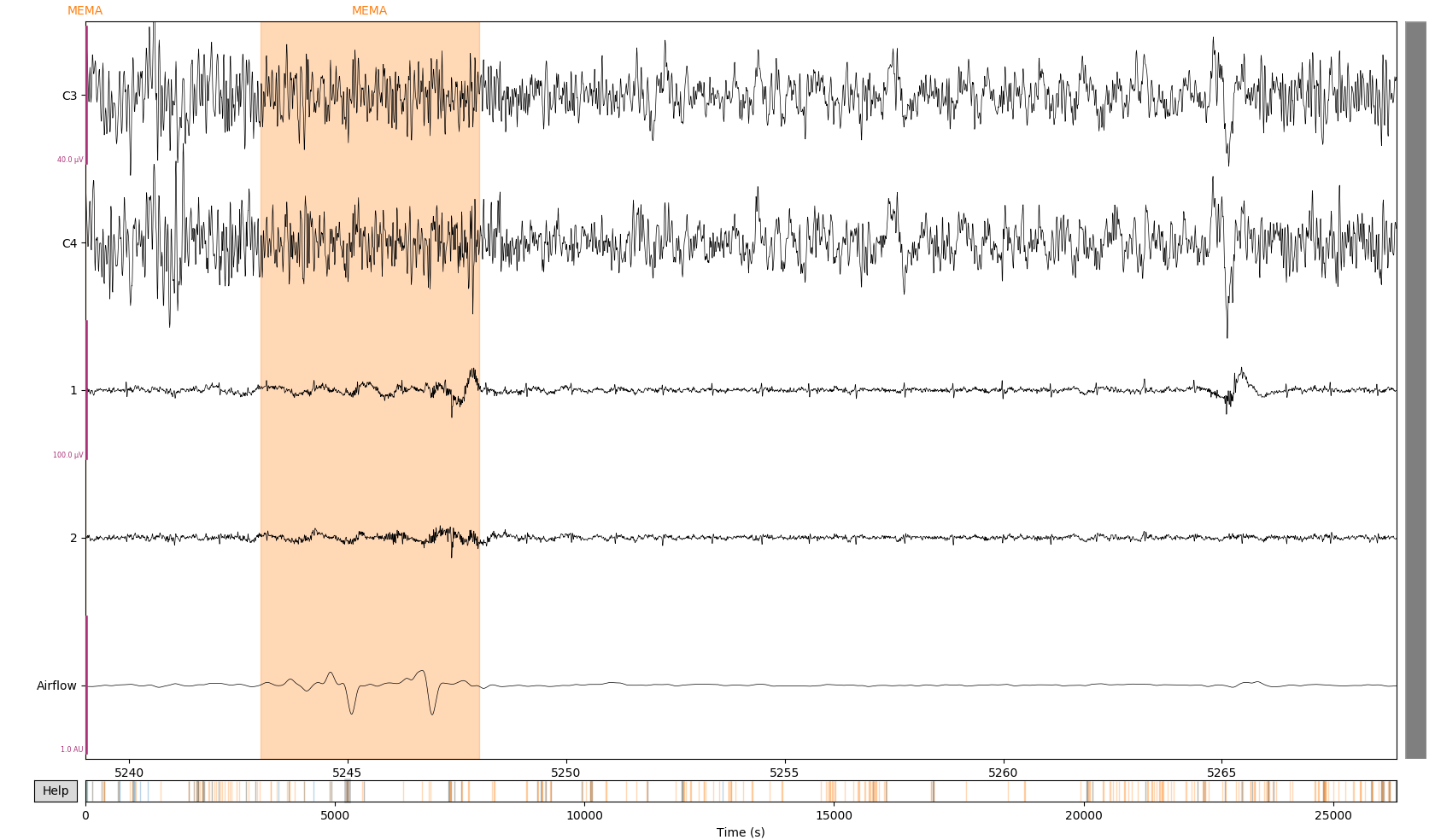 Figure 2: Detection of pure MEMA (orange)
Figure 2: Detection of pure MEMA (orange) episode consisting of two distant bursts. This two bursts are merged thank to the use of tinnsleep.events.scoring methods which allows to labels and categorize detected events.
There are several key parameters for classification (see Figure 3). We advice to used pre-configured AMDT pipeline using tinnsleep.pipeline.
CLASSIFICATION
- Window length: length of the sliding window for computer instantaneous power (default: 250ms for Bruxism, 1s for MEMA)
- Baseline: memory buffer for the computation of the baseline. By default, we advice to use the non-casual "left-hand"+"right-hand" baseline (see
tinnsleep.pipelinefor a pre-configured example) - Threshold: thresholding detects events for which power is X times greater than adaptive baseline
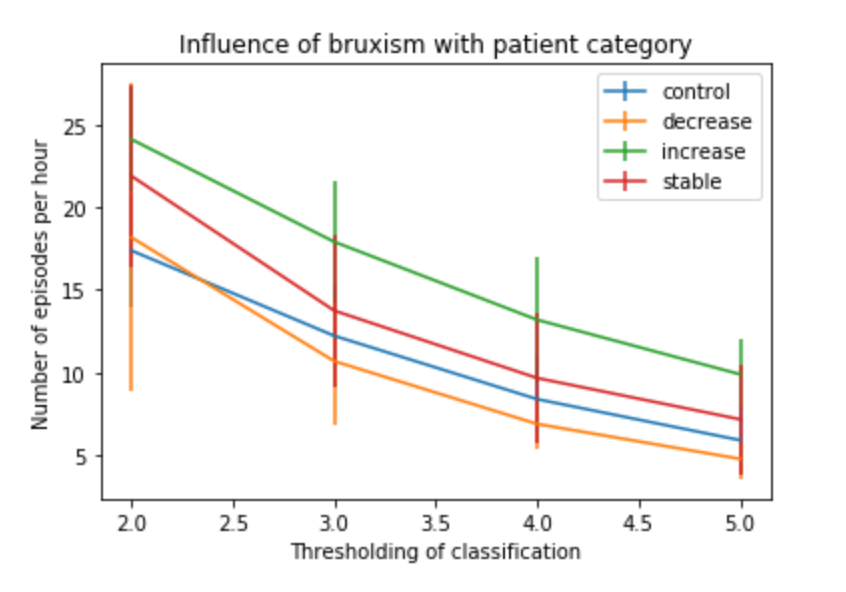 Figure 3: Influence of thresholding parameter in AMDT for the detection of the number of bruxism episodes per hour for each patient category (base on VAS-L).
Figure 3: Influence of thresholding parameter in AMDT for the detection of the number of bruxism episodes per hour for each patient category (base on VAS-L).
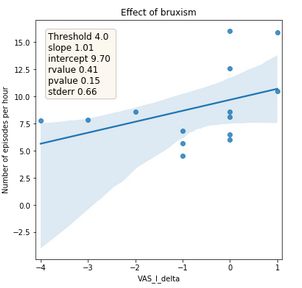
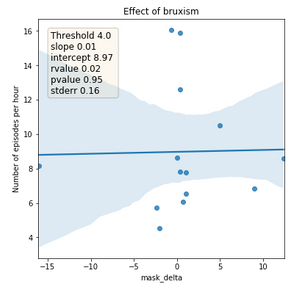 Figure 4: Relationship between the absolute difference of tinnitus masking volume between before sleep onset and after awakening (x-axis) and number of detected bruxism episodes per hour (y-axis). The line represents the trend and area the confidence interval.
Figure 4: Relationship between the absolute difference of tinnitus masking volume between before sleep onset and after awakening (x-axis) and number of detected bruxism episodes per hour (y-axis). The line represents the trend and area the confidence interval.
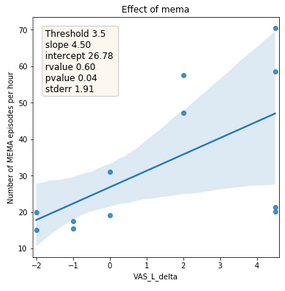
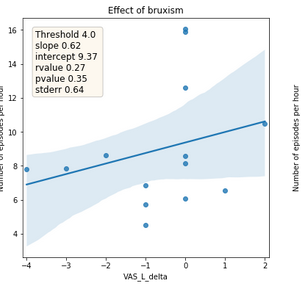 Figure 5: Relationship between the absolute difference of tinnitus subjective loudness between before sleep onset and after awakening (x-axis) and number of detected bruxism episodes per hour (y-axis). The line represents the trend and area the confidence interval.
Figure 5: Relationship between the absolute difference of tinnitus subjective loudness between before sleep onset and after awakening (x-axis) and number of detected bruxism episodes per hour (y-axis). The line represents the trend and area the confidence interval.
Figure 6: Relationship between the absolute difference of tinnitus subjective intrusiveness between before sleep onset and after awakening (x-axis) and number of detected bruxism episodes per hour (y-axis). The line represents the trend and area the confidence interval.
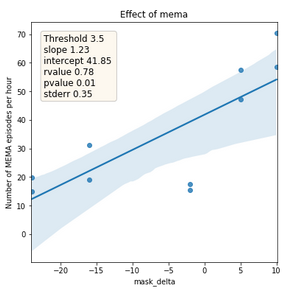
Figure 7: Relationship between the absolute difference of tinnitus masking volume between before sleep onset and after awakening (x-axis) and number of detected MEMA episodes per hour (y-axis). The line represents the trend and area the confidence interval.
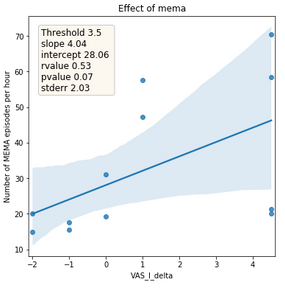
Figure 8: Relationship between the absolute difference of tinnitus subjective loudness between before sleep onset and after awakening (x-axis) and number of detected MEMA episodes per hour (y-axis). The line represents the trend and area the confidence interval.
Figure 9: Relationship between the absolute difference of tinnitus subjective intrusiveness volume between before sleep onset and after awakening (x-axis) and number of detected MEMA episodes per hour (y-axis). The line represents the trend and area the confidence interval.
The following steps must be performed on a Anaconda prompt console, or
alternatively, in a Windows command console that has executed the
C:\Anaconda3\Scripts\activate.bat command that initializes the PATH so that
the conda command is found.
-
Checkout this repository and change to the cloned directory for the following steps.
git clone git@github.com:lkorczowski/Tinnitus-n-Sleep.git cd Tinnitus-n-Sleep -
Create a virtual environment with all dependencies.
$ conda env create -f environment.yaml $ conda activate tinnsleep-env -
Activate the environment and install this package (optionally with the
-eflag).$ conda activate tinnsleep-env (tinnsleep-env)$ pip install -e . -
(optional) If you have a problem with a missing package, add it to the
environment.yaml, then:(tinnsleep-env)$ conda env update --file environment.yaml -
(optional) If you want to use the notebook, we advice Jupyter Lab (already in requirements) with additional steps:
$ conda activate tinnsleep-env # install jupyter lab (tinnsleep-env)$ conda install -c conda-forge jupyterlab (tinnsleep-env)$ ipython kernel install --user --name=tinnsleep-env (tinnsleep-env)$ jupyter lab # run jupyter lab and select tinnsleep-env kernel # quit jupyter lab with CTRL+C then (tinnsleep-env)$ conda install -c conda-forge ipympl (tinnsleep-env)$ conda install -c conda-forge nodejs (tinnsleep-env)$ jupyter labextension install @jupyter-widgets/jupyterlab-manager jupyter-matplotlibTo test if widget is working if fresh notebook:
import pandas as pd import matplotlib.pyplot as plt %matplotlib widget df = pd.DataFrame({'a': [1,2,3]}) plt.figure(2) plt.plot(df['a']) plt.show()
If you have trouble using git, a tutorial is available to describe :
- how to set git with github and ssh
- how to contribute, create a new repository, participate
- how to branch, how to pull-request
- several tips while using git
I love also the great git setup guideline written for BBCI Toolbox, just check it out!
A (incomplete) guideline is proposed to understand how to contribute and understand the best practices with branchs and Pull-Requests.