- Paper
- Software
- Axon Segmentation
- Brain Region Segmentation
- Brain Style Transfer
- Whole Brain Registration
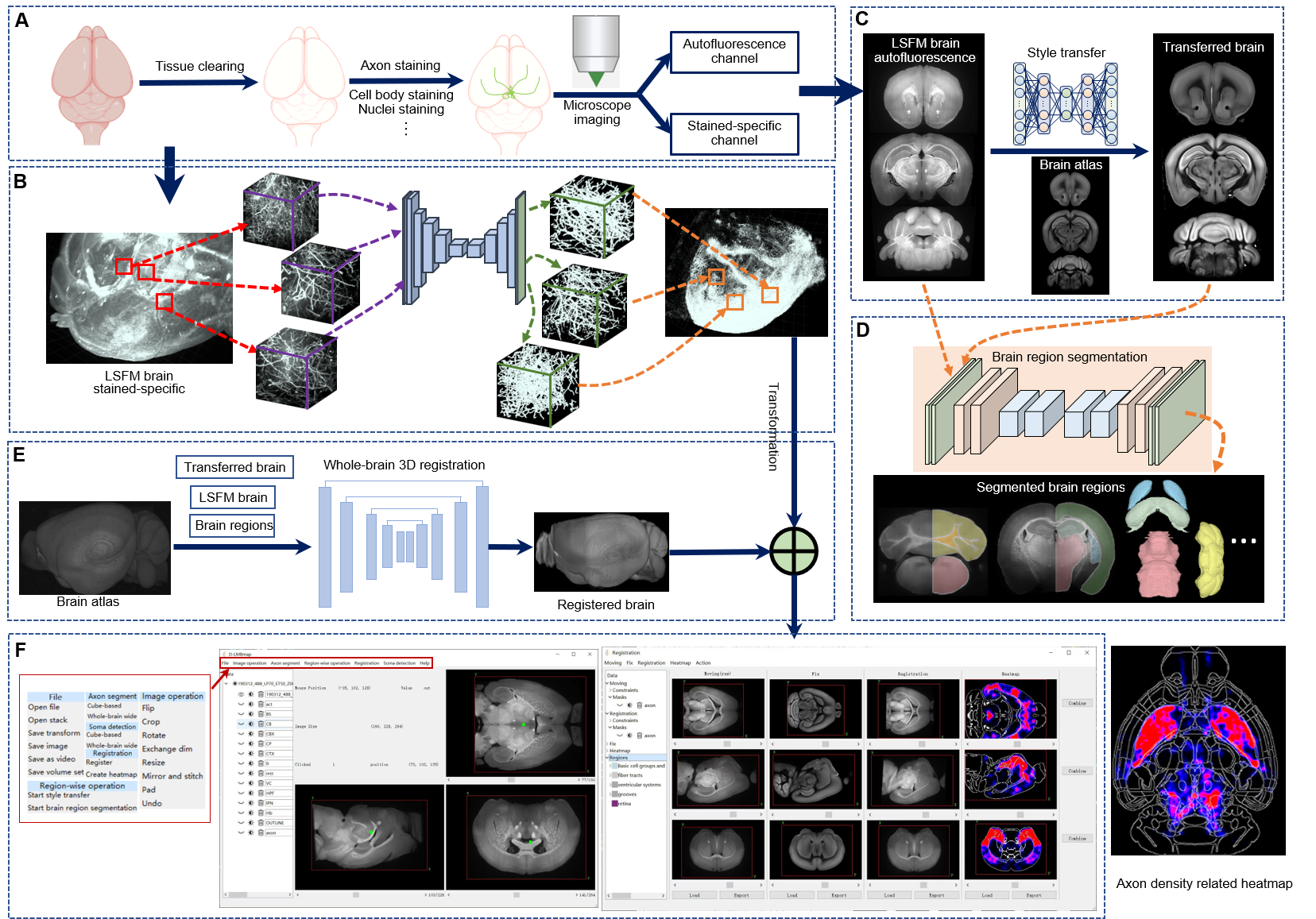
D-LMBmap, an end-to-end package providing an integrated workflow containing three modules based on novel deep-learning algorithms for whole-brain connectivity mapping: axon segmentation, brain region segmentation and whole-brain registration. D-LMBmap is the first method that does not require manual annotation for axon segmentation, and achieves quantitative analysis of whole-brain projectome in a single workflow with superior accuracy for multiple cell types in all of the modalities tested.
Our test dataset are accessible here.
D-LMBmap provides an open-source software with convenient high-level application programming interfaces (APIs) that can be used to build applications and extensions. The GUI provides functionality for the selection and computation of different pre-trained deep models. You can use D-LMBmap software by downloading our software from github->release.
Please refer to the tutorial before using the software.
 We provide a whole-brain axon segmentation pipeline and model training method, which can be accessed here.
We provide a whole-brain axon segmentation pipeline and model training method, which can be accessed here.
Axon segmentation models have been integrated into D-LMBmap software. The Axon segmentation function in the software can be used for cubes of any size, including whole-brain segmentation.
Brain region segmentation and brain style transfer methods mainly serve for whole brain registration. We also provide model training and prediction methods respectively, please refer to Brain style transfer and Brain region segmentation.
Some models have been integrated into D-LMBmap software.
 Here is the code for the multi-constraint and multi-scale facilitated whole-brain registration, axon density related heatmap generation.
Here is the code for the multi-constraint and multi-scale facilitated whole-brain registration, axon density related heatmap generation.
If you encounter any issues, please feel free to contact us at lipeiqi@stu.xjtu.edu.cn or zhongyuli@xjtu.edu.cn
This work is licensed under a Creative Commons Attribution 4.0 International License