small scripts useful for data wrangling at the LNCD
See the docs
Briefly clone and add to path
git clone https://github.com/lncd/lncdtools ~/lncdtools
echo "export PATH=\$PATH:$HOME/lncdtools" >> ~/.bashrc
- TODO: Makefile sentinels for
makeusingmkifdiff,mkls,mkmissing,mkstat - BIDS with
dcmdirtab,dcmtab_bids, andmknii
4dConcatSubBriks- extract a subbrick from a list of nifti label with luna ids. Useful for quality checking many structurals, subject masks, or individual contrasts. Wraps around 3dbucket and 3drefit:img_bg_rm- use imagemagick'sconvertto set a background to alpha (remove). Taken from "hackerb9" stack overflow solution. use on afni and suma screen capturesmkmissing- find missing patterns between two steps in a pipeline (file globs)r- read dataframe from stdin and run R code with shortcuts and magic a la DataScienceToolkit's Riotat2,melanin_align- modality specific wrappers
also see more detaied docs
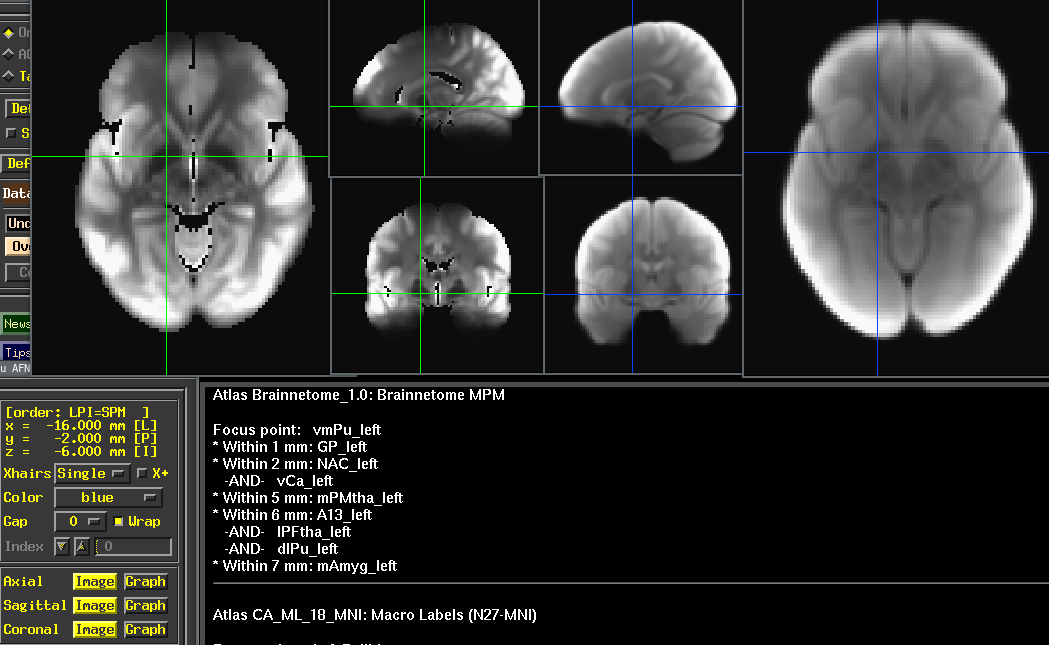 (
(/Volumes/Hera/Datasets/ABCD/TAT2/tat2_avg3797_med_voldisc.nii.gz and /Volumes/Hera/Projects/7TBrainMech/scripts/mri/tat2/mean_176.nii.gz)
permutation of tat2 calls were compared against R2 acquisitions:
-vol_median is likely the approprate normalization.
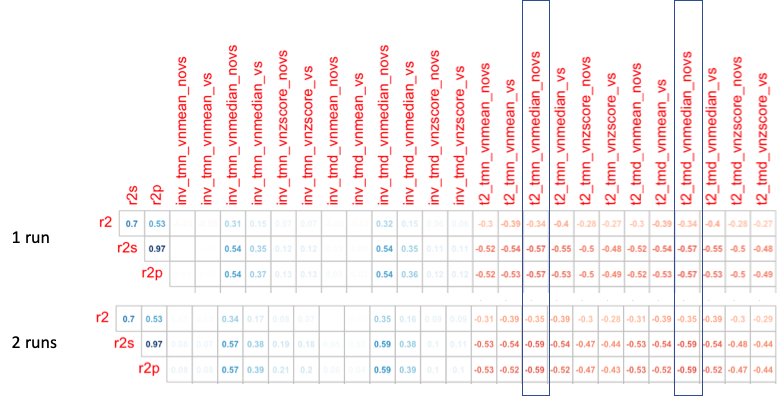
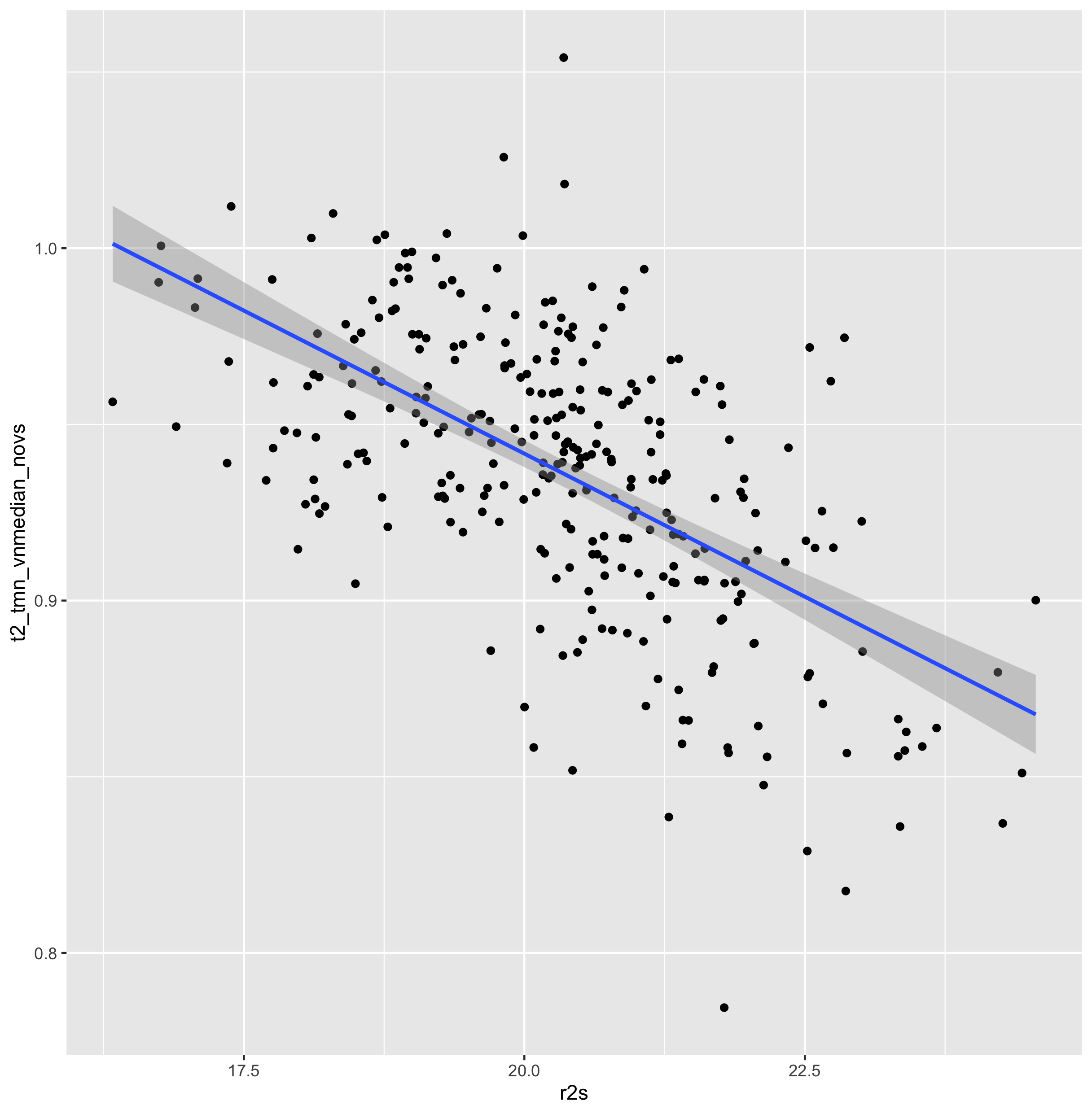
(/Volumes/Phillips/mMR_PETDA/scripts/tat2/multiverse)
get_ld8_age.Rrequires R and theLNCDRpackage + access with the firewall (for db atarnold.wpic.upmc.edu)