Mechanism of action and resistance to Trastuzumab Deruxtecan within the DAISY trial: machine learning analysis
Code used to carry on the machine learning analyses to study HER2 spatial distribution on baseline biopsy samples from the DAISY clinical trial.
To reproduce the analyses, please first start by indicating the correct paths in ./preprocessing/imports.py for:
SLIDE_PATHpointing towards raw whole slide images save location.ROIS_PATHpointing towards a csv file with ROI coordinates of areas to analyse in the slides.PATHOLOGIST_ANNOTATIONS_PATHpointing towards pathologist's annotations.
You also need the following csv files (considered to be located by default at the root of the code):
dataset.csvdescribing case ids, slide ids, cohort index, objective response to treatment and confirmation of objective response.correction_groups.csvdescribing the group attributed to each slide depending on the intensity of correction needed (only necessary for nuclei segmentation analysis).
See data access part for requiring access to these elements.
The packages needed to run the code are provided in environment.yml. The environment can be recreated using:
conda env create -f environment.yml
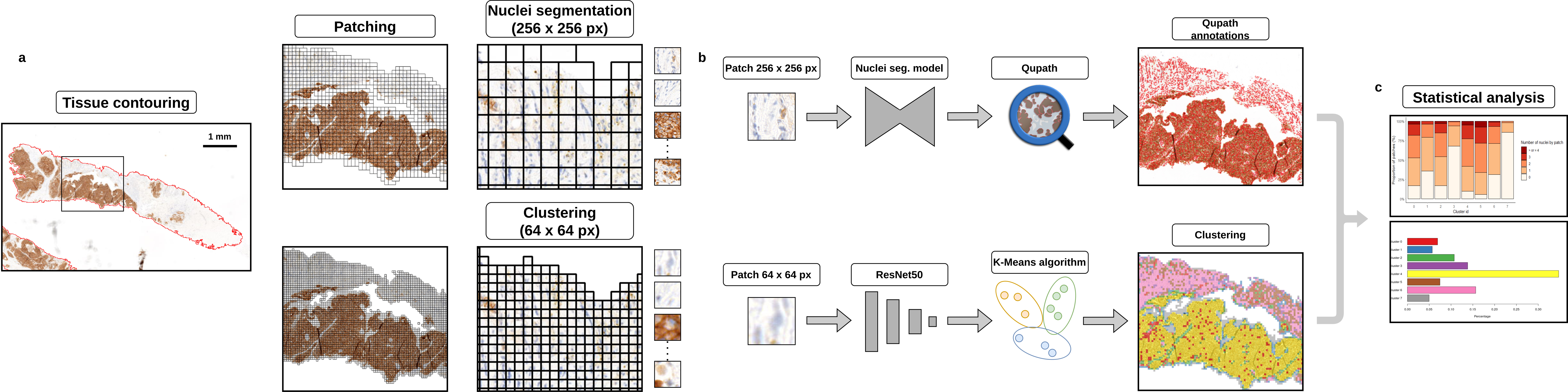
The slides are first pre-processed using ./create_patches_fp.py. To reproduce the clustering analysis, please use the following command line:
python ./create_patches_fp.py path/to/slides path/to/ROIs --pathologist_annotations path/to/pathologist/annotations --seg --patch --stitch --patch_size=64 --step_size=64 --save_dir ./Results/clustering/
To reproduce the nuclei segmentation analysis, please use the following command line:
python ./create_patches_fp.py path/to/slides path/to/ROIs --pathologist_annotations path/to/pathologist/annotations --seg --patch --stitch --patch_size=256 --step_size=128 --save_dir ./Results/nuclei/
For the clustering analysis, once the patches extracted, the ResNet features are computed using ./extract_features_fp.py with
the following command line:
python ./extract_features_fp.py --data_h5_dir ./Results/clustering/ --data_slide_dir path/to/slides --csv_path ./dataset.csv --feat_dir ./Results/clustering/feats --batch_size 512 --slide_ext .mrxs
Before performing clustering analysis, the extracted features must be concatenated in a matrix using ./compute_features_matrix with the following command line:
python ./compute_features_matrix.py --cohort_indices i
with i the index of the cohort to analyze.
Finally, the percentage of each cluster for each slide after training a Mini-Batch KMeans model on this feature matrix are retrieved using
./compute_cluster_percentages.py with the following command line:
python ./compute_cluster_percentages.py --features_matrix ./Results/clustering/features_matrix_cohort_i.pickle --cohort_indices i
with i the index of the cohort to analyze.
Optionally, the optimal number of clusters can be computed using ./optimal_number_clusters.
Several options are available to change the range of clusters to consider or score to use.
To reproduce the figures, please use the R script ./construct_figures_results.R, and the jupyter file visualize.ipynb.
Please feel free to contact us to get access to slide files, pathologist's annotations, and ROIs and dataset csv files.
The nuclei annotations are extracted using an unsupervised trained model available here (named model_daisy_HER2.pt). To reproduce the analysis, please use the following command line:
python ./script_extract_nuclei.py
You then need to import these annotations into QuPath and use the provided scripts in to extract nuclei features.
Lastly, the features are averaged across each patch used for clustering and compiled into a single csv file with clustering assignement. To reproduce the analysis, please use the following command line:
python ./script_compile_annotation_measurements.py --features_matrix ./Results/clustering/features_matrix_cohort_1.pickle --pretrained_kmeans ./Results/clustering/kmeans_model_cohort_1.pickle --cohort_indices 1
Many thanks to Lu et al. for providing the implementation of their whole slide images pre-processing on which this code is based on - see here for more details.
Please feel free to contact us for data access or any question at:
loic.le-bescond@gustaveroussy.fr
This code is MIT licensed, as found in the LICENSE file.