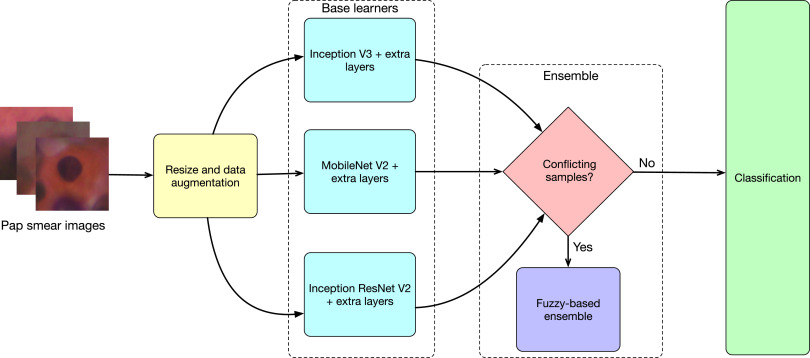
"A fuzzy distance-based ensemble of deep models for cervical cancer detection" published in Computer Methods and Programs in Biomedicine (June 2022), Elsevier
@article{pramanik2022fuzzy,
title = {A fuzzy distance-based ensemble of deep models for cervical cancer detection},
author={Pramanik, Rishav and Biswas, Momojit and Sen, Shibaprasad and de Souza J{\'u}nior, Luis Antonio and Papa, Jo{\~a}o Paulo and Sarkar, Ram},
journal = {Computer Methods and Programs in Biomedicine},
volume = {219},
pages = {106776},
year = {2022},
issn = {0169-2607},
doi = {10.1016/j.cmpb.2022.106776},
url = {https://www.sciencedirect.com/science/article/pii/S0169260722001626}
}
A fuzzy distance-based ensemble of deep models for cervical cancer detection
Find the original paper here.
Required directory structure:
(Note: train and test contains subfolders representing classes in the dataset.)
+-- data
| +-- train
| | +--class A
| | +--class B
| | ...
| +-- test
| | +--class A
| | +--class B
| | ...
+-- main.py
- Download the repository and install the required packages:
pip3 install -r requirements.txt
- The main file is sufficient to run the experiments. Then, run the code using linux terminal as follows:
python3 main.py --data_directory "data"
Available arguments:
--num_epochs: Number of epochs of training. Default = 70--learning_rate: Learning Rate. Default = 0.0001--batch_size: Batch Size. Default = 16--path: Data Path. Default= './'--kfold: K-Fold, to perform K fold cross validation. Default= 5
- Please don't forget to edit the above parameters before you start