Tensorflow implementation of the V-Net architecture for medical imaging segmentation.
This is a Tensorflow implementation of the V-Net architecture used for 3D medical imaging segmentation. This code adopts the tensorflow graph from https://github.com/MiguelMonteiro/VNet-Tensorflow. The repository covers training, evaluation and prediction modules for the (multimodal) 3D medical image segmentation in multiple classes.
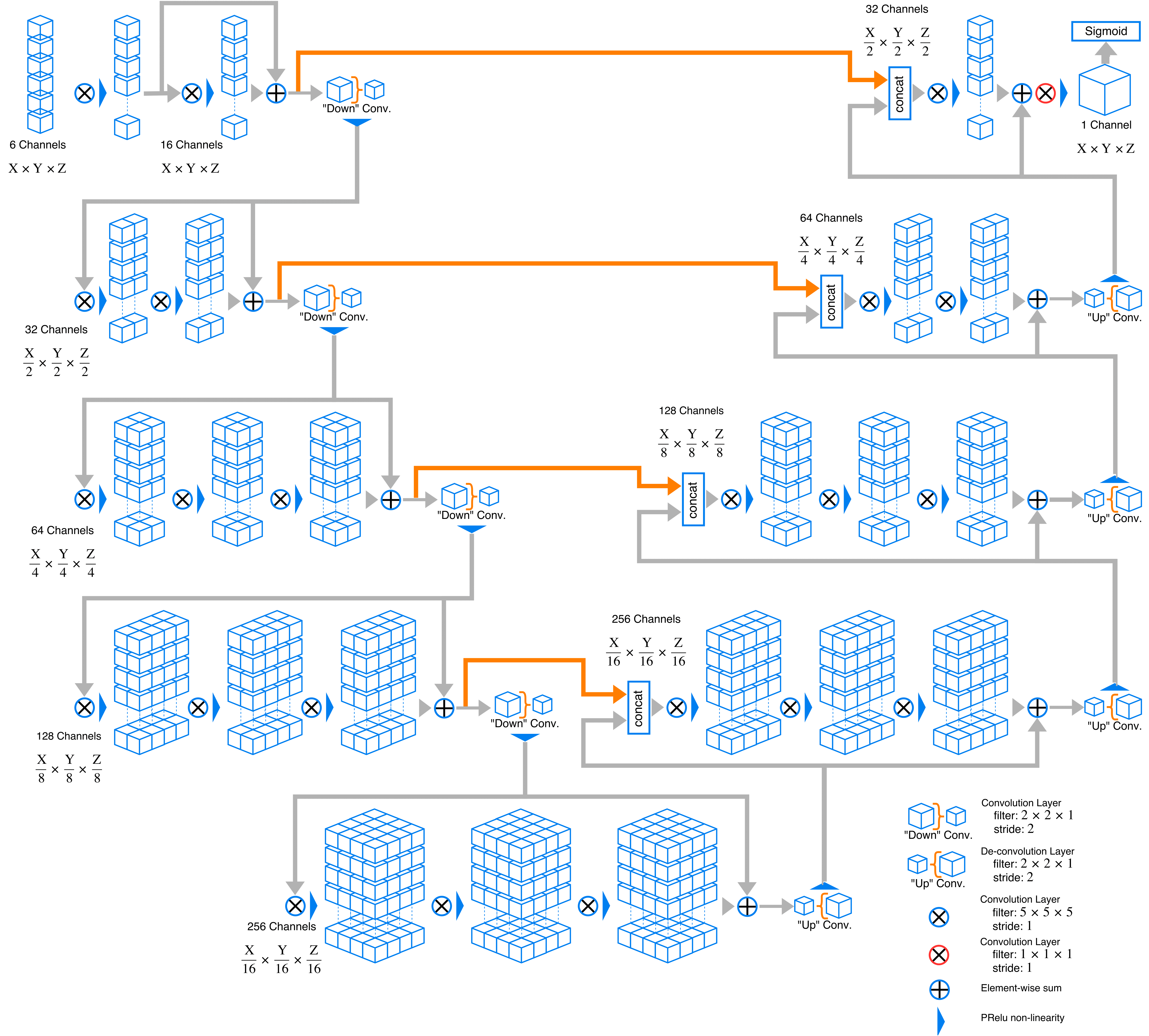
Here is an example graph of network this code implements. Channel depth may change owning to change in modality number and class number.
- 2D and 3D data processing ready
- Augmented patching technique, requires less image input for training
- Multichannel input and multiclass output
- Generic image reader with SimpleITK support (Currently only support .nii/.nii.gz format for convenience, easy to expand to DICOM, tiff and jpg format)
- Medical image pre-post processing with SimpleITK filters
- Easy network replacement structure
- Sørensen and Jaccard similarity measurement as golden standard in medical image segmentation benchmarking
- Utilizing medical image headers to retrive space and orientation info after passthrough the network
- Training
- Tensorboard visualization and logging
- Resume training from checkpoint
- Epoch training
- Evaluation from single data
- Multichannel input
- Multiclass output
- Weighted DICE loss
- C++ inference (Deprecated)
Known good dependencies
- Python 3.7
- Tensorflow 1.14 or above
- SimpleITK
All training, testing and evaluation data should put in ./data
.
├── ...
├── data # All data
│ ├── testing # Put all testing data here
| | ├── case1
| | | ├── img.nii.gz # Image for testing
| | | └── label.nii.gz # Corresponding label for testing
| | ├── case2
| | ├──...
│ ├── training # Put all training data here
| | ├── case1 # foldername for the cases is arbitary
| | | ├── img.nii.gz # Image for training
| | | └── label.nii.gz # Corresponding label for training
| | ├── case2
| | ├──...
│ └── evaluation # Put all evaluation data here
| | ├── case1 # foldername for the cases is arbitary
| | | └── img.nii.gz # Image for evaluation
| | ├── case2
| | ├──...
├── tmp
| ├── cktp # Tensorflow checkpoints
| └── log # Tensorboard logging folder
├── ...
If you wish to use image and label with filename other than img.nii.gz and label.nii.gz, please change the following values in config.json
"ImageFilenames": ["img.nii.gz"],
"LabelFilename": "label.nii.gz"The network will automatically select 2D/3D mode by the length of PatchShape in config.json
In segmentation tasks, image and label are always in pair, missing either one would terminate the training process.
The code has been tested with LiTS dataset
You may run train.py with commandline arguments. To check usage, type python main.py -h in terminal to list all possible training parameters.
Available training parameters
-h, --help show this help message and exit
-v, --verbose Show verbose output
-p [train evaluate], --phase [train evaluate]
Training phase (default= train)
--config_json FILENAME
JSON file for model configuration
--gpu GPU_IDs Select GPU device(s) (default = 0)The program will read the configuration from config.json. Modify the necessary hyperparameters to suit your dataset.
Note: You should always set label 0 as the first SegmentationClasses in config.json. Current model will only run properly with at least 2 classes.
The software will automatically determine run in 2D or 3D mode according to rank of PatchShape in config.json
Typically medical image is large in size when comparing with natural images (height x width x layers x modality), where number of layers could up to hundred or thousands of slices. Also medical images are not bounded to unsigned char pixel type but accepts short, double or even float pixel type. This will consume large amount of GPU memories, which is a great barrier limiting the application of neural network in medical field.
Here we introduce serveral data augmentation skills that allow users to normalize and resample medical images in 3D sense. In train.py, you can access to trainTransforms/testTransforms. For general purpose we combine the advantage of tensorflow dataset api and SimpleITK (SITK) image processing toolkit together. Following is the preprocessing pipeline in SITK side to facilitate image augmentation with limited available memories.
- Image Normalization (fit to 0-255)
- Isotropic Resampling (adjustable size, in mm)
- Padding (allow input image batch smaller than network input size to be trained)
- Random Crop (randomly select a zone in the 3D medical image in exact size as network input)
- Gaussian Noise
The preprocessing pipeline can easily be adjusted with following example code in train.py:
trainTransforms = [
NiftiDataset3D.Normalization(),
NiftiDataset3D.Resample(self.spacing[0],self.spacing[1],self.spacing[2]),
NiftiDataset3D.Padding((self.patch_shape[0], self.patch_shape[1], self.patch_shape[2])),
NiftiDataset3D.RandomCrop((self.patch_shape[0], self.patch_shape[1], self.patch_shape[2]),self.drop_ratio,self.min_pixel),
NiftiDataset3D.RandomNoise()
]For 2D image training mode, you need to provide transforms in 2D and 3D separately.
To write you own preprocessing pipeline, you need to modify the preprocessing classes in NiftiDataset.py
Additional preprocessing classes:
-
StatisticalNormalization
-
Reorient (take care on the direction when performing evaluation)
-
Invert
-
ConfidenceCrop (for very small volume like cerebral microbleeds, alternative of RandomCrop)
-
Deformations: The deformations are following SITK deep learning data augmentation documentations, will be expand soon. Now contains:
- BSplineDeformation
Hint: Directly apply deformation is slow. Instead you can first perform cropping with a larger than patch size region then with deformation, then crop to actual patch size. If you apply deformation to exact training size region, it will create black zone which may affect final training accuracy.
Example:
NiftiDataset.ConfidenceCrop((self.patch_shape[0]*2, self.patch_shape[1]*2, self.patch_shape[2]*2),(0.0001,0.0001,0.0001)), NiftiDataset.BSplineDeformation(), NiftiDataset.ConfidenceCrop((self.patch_shape[0], self.patch_shape[1], self.patch_shape[2]),(0.5,0.5,0.25)),
In training stage, result can be visualized via Tensorboard. Run the following command:
tensorboard --logdir=./tmp/logOnce TensorBoard is running, navigate your web browser to localhost:6006 to view the TensorBoard.
Note: localhost may need to change to localhost name by your own in newer version of Tensorboard.
To evaluate image data, first place the data in folder ./data/evaluate. Each image data should be placed in separate folder as indicated in the folder hierarchy
There are several parameters you need to set in order manually
model_path, the default path is at./tmp/ckpt/checkpoint-<global_step>.metacheckpoint_dir, the default path is at./tmp/ckptpatch_size, this value need to be same as the one used in trainingpatch_layer, this value need to be same as the one used in trainingstride_inplane, this value should be <=patch_sizestride_layer, this value should be <=patch_layerbatch_size, currently only support single batch processing
Run evaluate.py after you have modified the corresponding variables. All data in ./data/evaluate will be iterated. Segmented label is named as label_vnet.nii.gz in same folder of the respective img.nii.gz.
You may change output label name by changing the line writer.SetFileName(os.path.join(FLAGS.data_dir,case,'label_vnet.nii.gz'))
Note that you should keep preprocessing pipeline similar to the one in train.py, but without random cropping and noise.
We provide a C++ inference example under directory cxx. For C++ implementation, please follow the guide here
Use the following Bibtex if you need to cite this repository:
@misc{jackyko1991_vnet_tensorflow,
author = {Jacky KL Ko},
title = {Implementation of vnet in tensorflow for medical image segmentation},
howpublished = {\url{https://github.com/jackyko1991/vnet-tensorflow}},
year = {2018},
publisher={Github},
journal={GitHub repository},
}
@inproceedings{milletari2016v,
title={V-net: Fully convolutional neural networks for volumetric medical image segmentation},
author={Milletari, Fausto and Navab, Nassir and Ahmadi, Seyed-Ahmad},
booktitle={3D Vision (3DV), 2016 Fourth International Conference on},
pages={565--571},
year={2016},
organization={IEEE}
}
@misc{MiguelMonteiro_VNet_Tensorflow,
author = {Miguel Monteiro},
title = {VNet-Tensorflow: Tensorflow implementation of the V-Net architecture for medical imaging segmentation.},
howpublished = {\url{https://github.com/MiguelMonteiro/VNet-Tensorflow}},
year = {2018},
publisher={Github},
journal={GitHub repository},
}- SimpleITK guide on deep learning data augmentation: https://simpleitk.readthedocs.io/en/master/Documentation/docs/source/fundamentalConcepts.html https://simpleitk.github.io/SPIE2018_COURSE/data_augmentation.pdf https://simpleitk.github.io/SPIE2018_COURSE/spatial_transformations.pdf
Jacky Ko jackkykokoko@gmail.com