SequenceTools is a package for processing and analysing biological sequence alignments, with a focus is on protein.
# Currently the only way to install it is over github
install.packages("devtools")
devtools::install_github("ltschmitt/SequenceTools")You can read in fasta files of DNA or amino acid single letter using read_alignments. The output is a named character vector, which makes further processing uncomplicated. An example how to read in a fasta file, generate a consensus sequence and to plot the alignment of all the reads.
library(SequenceTools)
library(tidyverse, verbose = F)
# read in single line fasta
seqs = read_alignments(input = 'data_raw/cre-variants.fa', naming = 'headers')
#> reading 1 sequence file(s)...
# make consensus sequence
# sequences need to be same length
# consensus is made with the sequences of the same name
consens = generate_consensus(setNames(seqs, rep('consens',length(seqs))))
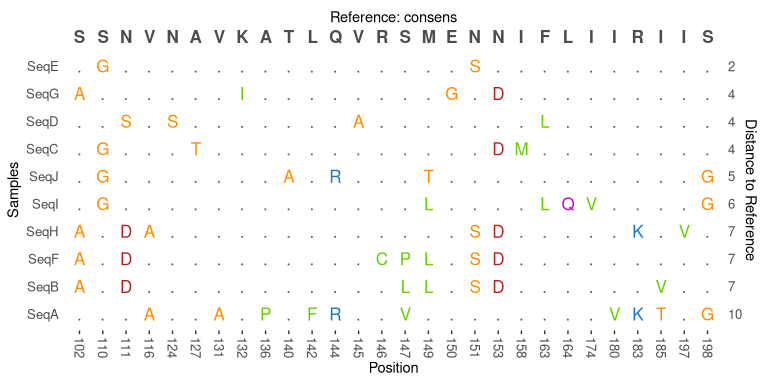
# plot alignment with consens as reference, makes ggplot element
plot_alignment(Alignment = seqs, Reference = consens, seqrange = 100:200) +
theme(legend.position = 'none')# convert the sequences to a "long" format which makes working with tidyverse tools very easy
seqs = read_alignments(input = 'data_raw/cre-variants.fa', naming = 'filenames')
#> reading 1 sequence file(s)...
lseqs = alignments2long(seqs, get_frequencies = T)
head(lseqs)
#> # A tibble: 6 × 5
#> Sample Pos AA n Freq
#> <fct> <int> <chr> <int> <dbl>
#> 1 cre-variants 1 M 10 1
#> 2 cre-variants 2 A 1 0.1
#> 3 cre-variants 2 S 6 0.6
#> 4 cre-variants 2 T 3 0.3
#> 5 cre-variants 3 D 5 0.5
#> 6 cre-variants 3 N 5 0.5
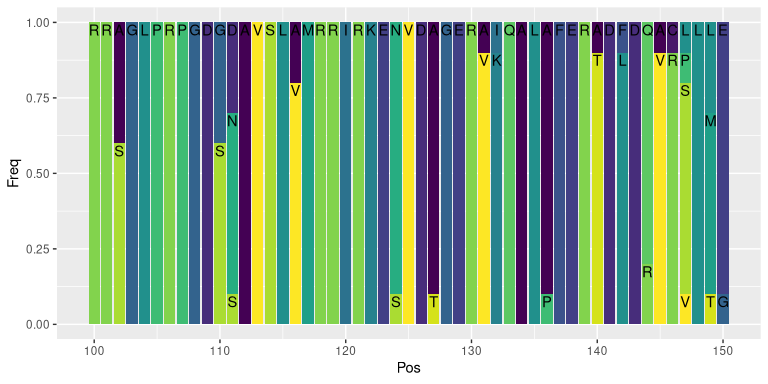
lseqs %>%
filter(Pos %in% 100:150) %>%
ggplot(aes(x = Pos, y = Freq, fill = AA, label = AA)) +
geom_col() +
geom_text(position = 'stack', vjust = 1.2) +
scale_fill_viridis_d() +
theme(legend.position = 'none')- aa_properties: converts single letter amino acid sequences into the requested property
- alignment_heatmap: makes a heatmap of an alignment with colors indicating amino acids.
- alignment_tsne: makes a tsne dimensionality reduction of an alignment
- alignments2long: converts the alignment into a long format with sequence position and single AA/DNA letters.
- between_lib_distances: plots the all-vs-all hamming distances between two libraries
- color_switcher: takes a color pallette and mixes the color order
- generate_consensus: generates the consensus sequences of an alignment
- gg_color_hue: emulates the standard ggplot color pallette
- hamming_heatmap: makes a heatmap of an alignment according to all-vs-all hamming distances
- plot_alignment: plots an alignment
- plot_frequencies: plots the frequences of the amino acids in the library
- read_alignments: read in alignments as named character vector
- reverse_complement: converts DNA sequence into the reverse complement, works with IUPAC bases
- within_lib_distances: plots the all-vs-all hamming distances within the libraries
- write_fasta: write sequences into a fasta file format
Since this package was developed for my own use, it might contain minor issues and is not fully developed/consistent. Feel free to let me know, but I don’t guarantee that I will do something about it.
This package was developed to suite my preferences, your might want something different. Here are some other R packages I found to be useful with similar functions: