UDA is a collection of methods concerning unsupervised domain adaptation techniques, developed for the Deep Learning course of the master's degree program in Computer Science at the University of Trento.
The dataset employed is the Adaptiope unsupervised domain adaptation benchmark, considering only few classes and only two domains for the sake of simplicity of the project.
To simplify the task the domains considered are product and real world whose chosen categories are: backpack, bookcase, car jack, comb, crown, file cabinet, flat iron, game controller, glasses, helicopter, ice skates, letter tray, monitor, mug, network switch, over-ear headphones, pen, purse, stand mixer and stroller.
Here we link the wandb experiments' learning curves and the Jupyter notebook.
Note: more information can be found in the
report
| Name | Surname | Username |
|---|---|---|
| Samuele | Bortolotti | samuelebortolotti |
| Luca | De Menego | lucademenego99 |
The objective of the repository is to devise, train and evaluate a deep learning framework on an Unsupervised Domain Adaptation setting.
The test dataset on which the proposed methods are tested is the Adaptiope, a large scale, diverse UDA dataset with 3 domains: synthetic, product and real world data.
The implemented networks will be trained:
- in a supervised way on the source domain;
- in an unsupervised way on the target domain.
The gain, namely the amount of improvement we receive when we use the domain adaption framework compared to the outcome we get without any domain alignment approach, is used to assess the quality of the domain alignment strategy. The gain can be represented mathematically as:
This has been be done in both directions between the two selected domains, using as evaluation metric the validation accuracy on the target domain:
Where:
-
$TP$ stands for true positives, namely cases in which the algorithm correctly predicted the positive class -
$TN$ stands for true negatives, namely cases in which the algorithm correctly predicted the negative class -
$FP$ stands for false positives, namely cases in which samples are associated with the positive class whereas they belong to the negative one. (Also known as a "Type I error.") -
$FN$ stands for false negatives, namely cases in which samples are associated with the negative class whereas they belong to the positive one. (Also known as a "Type II error.")
Eventually lower and upper bounds will be calculated training the networks only on one specific domain (source domain for alower bound, target domain for an upper bound), enabling us to provide some considerations about the achievements of our architectures.
Just to give a formal background, we leverage on the definition of Domain Adaptation provided by Gabriela Csurka et.al which in the following sub-section will be presented.
We define a domain
Given sample set
Let us assume we have two domains with their related tasks: a source domain
If the two domains corresponds, namely
However, this assumption is violated,
Domain Adaptation, usually refers to problems in which the tasks are considered to be the same (
In a classification task, both the set of labels and the conditional distributions are assumed to be shared between the two domains, i.e.
The second assumption,
Furthermore, the Domain Adaptation community distinguishes between the unsupervised scenario, in which labels are only accessible for the source domain, and the semi-supervised condition, in which only a limited number of target samples are labeled.
When dealing with Deep Domain Adaptation problems, there are three main ways to solve them:
- discrepancy-based methods: align domain data representation with statistical measures;
- adversarial-based methods: involve also a domain discriminator to enforce domain confusion;
- reconstruction-based methods: use an auxiliary reconstruction task to ensure a domain-invariant feature representation.
The repository contains a possible solution for each one of the methods depicted above, together with a customized solution which combines two of them: discrepancy-based and reconstruction-based (DSN + MEDM).
Having only few examples available, training a network from scratch has not given satisfactory results; that is the reason why we have decided to always start with a feature extractor pre-trained on the ImageNet object recognition dataset.
Moreover, in order to delve into the proposed methods, we have given the opportunity to evaluate the domain adaptation method starting from two different backbones networks, namely AlexNet and ResNet18.
To successfully meet the domain adaptation challenge, we have implemented several network architectures which are then adopted from relevant papers in the literature and altered to fit our needs.
Our major goal, as stated in the introduction, is to evaluate the performance (gain) of some of the most significant approaches in the domain adaptation field and build them on top of two of the most relevant convolutional neural network architectures, namely AlexNet and ResNet.
In this repository we cover:
- Deep Domain Confusion Networks
- Domain Adversarial Neural Network
- Domain Separation Networks
- Rotation loss Network
- Entropy Minimization vs. Diversity Maximization for Domain Adaptation
- A trial of ours to combine
MEDMwithDANN - Another idea of ours to combine
MEDMwithDSN
In this project we have tried out at least one architecture for each domain adaptation category of techniques, namely discrepancy-based, adversarial-based and reconstruction-based. Among them, the most effective ones were DANN and MEDM.
MEDM is an approach that can be easily integrated in any architecture, since it consists of two simple entropy losses, showing a stable training and astonishing performance. On the other hand, DANN is much more subject to hyperparameter variations and it is very likely to end up in an unstable training.
In our testing, we found that DSN and MEDM worked effectively together, whereas MEDM and DANN combined have not given a consistent improvement.
Since it's more difficult to adapt from product to real world, gain is significantly simpler to obtain because there is far more space for improvement between the baseline and upper bound.
On the other hand, switching between real world and product images guarantees higher accuracy because networks have already seen real world images since they were pre-trained on ImageNet.
As a result, the gap between the baseline and the real world is significantly narrower, which means getting a significant progress is much harder.
After having tried out both AlexNet and ResNet18 as backbone architectures, we found that not only did ResNet18 have far higher baseline and upper-bound values, but also all domain adaption strategies produced significantly larger gains.
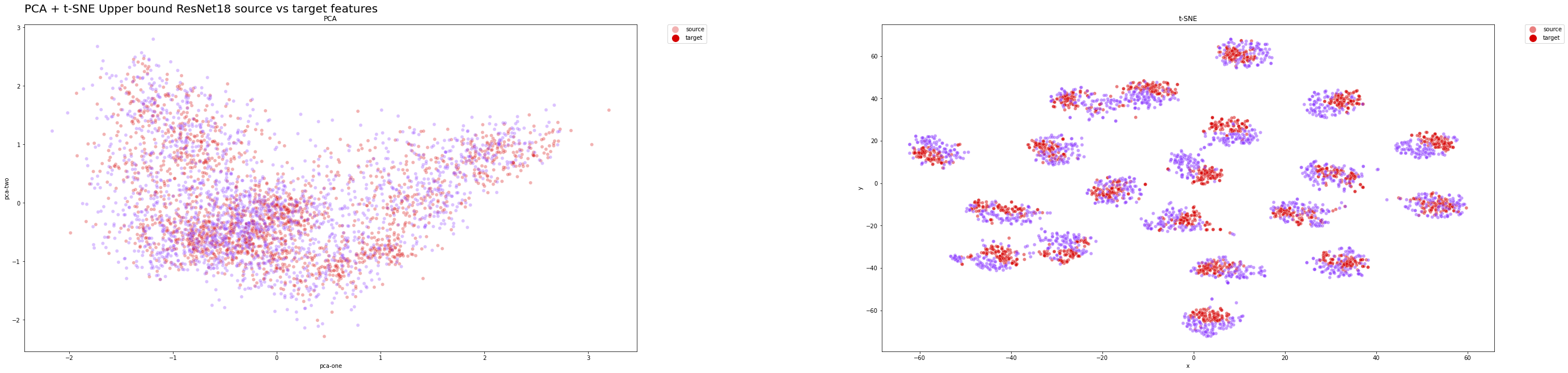
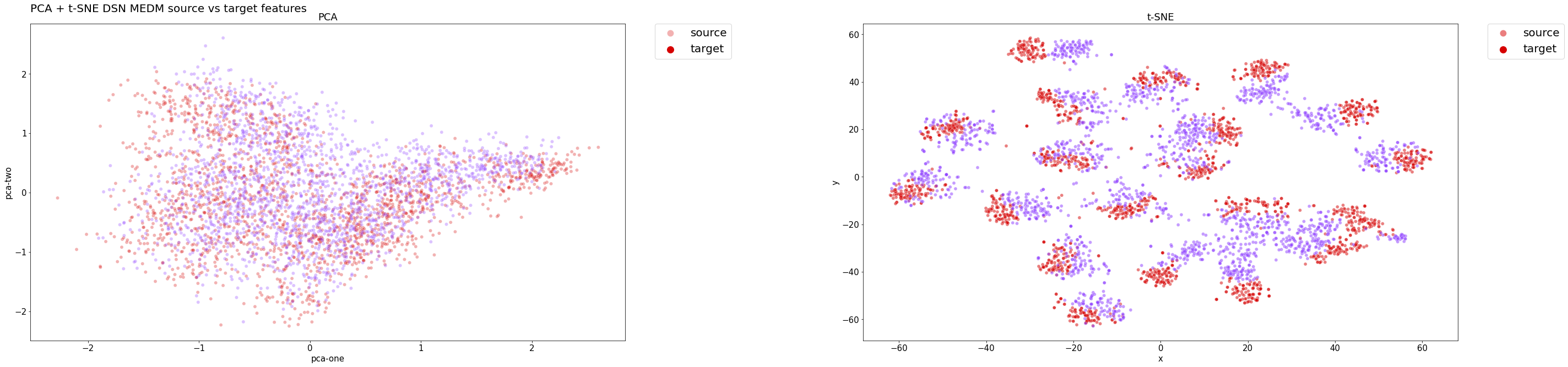
The t-SNE plots has clearly shown our last solution is able to extract relevant features from both domains while performing an accurate classification, since the distributions are close to the optimal ones, which are those of the upper bound ResNet18 model.
We have provided some plots to observe how the features of target and source domain are distributed thanks to dimensionality reduction techniques.
Here, for instance, we have shown the product-to-real-world domain adaptation, considering only the model that has achieved the best performance, namely DSN + MEDM. So as to give an effective representation of the distribution alignment and provide a single graph, which is the most significant one regarding our project.
In the following sections two graphs are presented:
- one depicting the distribution of the features learned by backbone neural network (ResNet18 in both cases) without considering the classifier or bottleneck layers.
- one portraying the predicted class distribution.
Note: that in both cases the plots are computed on the target domain (
real-world), on which theDSN+MEDMapproach has not access.
Ideally, we would like that our domain adaptation model achieves the performances of the upper-bound model, which is the one that best generalize the source and target domain samples.
Therefore, the model feature distribution is a good method to assess the quality of the domain adaptation technique.
Upper bound features distribution on the test set:
Upper bound classes distribution on the test set:
We expect that both the classes and the features distribution are arranged in a similar way with respect to the Target distributions, as the model has achieved astonishing performances.
DSN + MEDM features distribution on the test set:
As we can see from the classes distributions below, even the model's classifier is able to effectively cluster the examples while maintaining a high accuracy. Clusters are well defined and separated between them. Despite being examples of the target domain, they are correctly classified.
DSN + MEDM classes distribution on the test set:
In both cases, PCA hasn't really provided useful insights about the results, since a linear transformation may be too naive to transform such high dimensional data into a lower dimensional embedding.
Here we provide the results we have achieved on the smaller version of Adaptiope:
| Backbone Network | Method | Source set | Target set | Accuracy on target (%) | Gain |
|---|---|---|---|---|---|
| AlexNet | Source Only | Product Images | Real Life | 64.45 | 0 |
| ResNet18 | Source Only | Product Images | Real Life | 75.39 | 0 |
| AlexNet | Upper Bound | Product Images | Real Life | 85.94 | 21.485 |
| ResNet18 | Upper Bound | Product Images | Real Life | 96.88 | 20.312 |
| AlexNet | DDC | Product Images | Real Life | 68.36 | 3.91 |
| ResNet18 | DDC | Product Images | Real Life | 83.50 | 8.11 |
| AlexNet | DANN | Product Images | Real Life | 65.23 | 0.78 |
| ResNet18 | DANN | Product Images | Real Life | 87.11 | 11.72 |
| OriginalDSN | Product Images | Real Life | 04.17 | ||
| AlexNet | BackboneDSN | Product Images | Real Life | 58.85 | -5.60 |
| ResNet18 | BackboneDSN | Product Images | Real Life | 78.39 | 3.00 |
| ResNet18 | OptimizedDSN | Product Images | Real Life | 81.25 | 5.86 |
| ResNet18 | Rotation Loss | Product Images | Real Life | 73.83 | -1.56 |
| ResNet18 | MEDM | Product Images | Real Life | 90.36 | 14.97 |
| ResNet18 | MEDM + DANN | Product Images | Real Life | 80.21 | 4.82 |
| ResNet18 | MEDM + DSN | Product Images | Real Life | 91.15 | 15.76 |
| AlexNet | Source Only | Real Life | Product Images | 85.55 | 0 |
| ResNet18 | Source Only | Real Life | Product Images | 93.75 | 0 |
| AlexNet | Upper Bound | Real Life | Product Images | 95.70 | 10.15 |
| ResNet18 | Upper Bound | Real Life | Product Images | 98.05 | 4.3 |
| AlexNet | DDC | Real Life | Product Images | 82.03 | -3.52 |
| ResNet18 | DDC | Real Life | Product Images | 94.53 | 0.78 |
| AlexNet | DANN | Real Life | Product Images | 83.98 | -1.57 |
| ResNet18 | DANN | Real Life | Product Images | 96.09 | 2.34 |
| OriginalDSN | Real Life | Product Images | 04.17 | ||
| AlexNet | BackboneDSN | Real Life | Product Images | 84.11 | -1.44 |
| ResNet18 | BackboneDSN | Real Life | Product Images | 91.41 | -2.34 |
| ResNet18 | OptimizedDSN | Real Life | Product Images | 93.49 | -0.26 |
| ResNet18 | Rotation Loss | Real Life | Product Images | 91.80 | -1.95 |
| ResNet18 | MEDM | Real Life | Product Images | 97.14 | 3.39 |
| ResNet18 | MEDM + DANN | Real Life | Product Images | 94.00 | 0.25 |
| ResNet18 | MEDM + DSN | Real Life | Product Images | 96.61 | 2.86 |
The code as-is runs in Python 3.7 with the following dependencies
And the following development dependencies
Follow these instructions to set up the project on your PC.
Moreover, to facilitate the use of the application, a Makefile has been provided; to see its functions, simply call the appropriate help command with GNU/Make
make helpgit clone https://github.com/samuelebortolotti/uda.git
cd udapip install --upgrade pip
pip install -r requirements.txtNote: it might be convenient to create a virtual enviroment to handle the dependencies.
The
Makefileprovides a simple and convenient way to manage Python virtual environments (see venv). In order to create the virtual enviroment and install the requirements be sure you have the Python 3.7 (it should work even with more recent versions, however I have tested it only with 3.7)make env source ./venv/uda/bin/activate make installRemember to deactivate the virtual enviroment once you have finished dealing with the project
deactivate
The automatic code documentation is provided Sphinx v4.5.0.
In order to have the code documentation available, you need to install the development requirements
pip install --upgrade pip
pip install -r requirements.dev.txtSince Sphinx commands are quite verbose, I suggest you to employ the following commands using the Makefile.
make doc-layout
make docThe generated documentation will be accessible by opening docs/build/html/index.html in your browser, or equivalently by running
make open-docHowever, for the sake of completness one may want to run the full Sphinx commands listed here.
sphinx-quickstart docs --sep --no-batchfile --project unsupervised-domain-adaptation --author "Samuele Bortolotti, Luca De Menego" -r 0.1 --language en --extensions sphinx.ext.autodoc --extensions sphinx.ext.napoleon --extensions sphinx.ext.viewcode --extensions myst_parser
sphinx-apidoc -P -o docs/source .
cd docs; make htmlNote: executing the second list of command will lead to a slightly different documentation with respect to the one generated by the
Makefile. This is because the above listed commands do not customize the index file of Sphinx. This is because the above listed commands do not customise the index file of Sphinx.
To prepare the Adaptiope dataset filtered out, you can type:
python -m uda dataset [--classes ADDITIONAL_CLASSES --destination-location DATASET_DESTINATION_LOCATION --new-dataset-name FILTERED_DATASET_NAME]where classes is the list of additional classes to filter out, by default they are:
classes = [
"backpack",
"bookcase",
"car jack",
"comb",
"crown",
"file cabinet",
"flat iron",
"game controller",
"glasses",
"helicopter",
"ice skates",
"letter tray",
"monitor",
"mug",
"network switch",
"over-ear headphones",
"pen",
"purse",
"stand mixer",
"stroller",
]destination_location refers to the name of the zip folder of the Adaptiope dataset which will be downloaded automatically by the script; whereas new-dataset-name is the name
of the new filtered dataset folder.
Alternatively, you can obtain the same result in a less verbose manner by tuning the flags in the Makefile and then run:
make datasetTo have a simple overview of how a label in the dataset looks like you can type:
python -m uda visualize DATASET_SOURCE DATASET_DEST LABEL [--batch-size BATCH_SIZE]where DATASET_SOURCE is the source dataset folder, DATASET_DEST is the destination dataset location and LABEL is the label of the data you want the script to display.
As an effective representation the command:
python -m uda visualize adaptiope_small/product_images adaptiope_small/real_life 8Alternatively, you can obtain the same result in a less verbose manner by tuning the flags in the Makefile and then run:
make visualizeNote: by default the
adaptiope_small/product_images,adaptiope_small/real_lifeand8respectively asSOURCE_DATASET,TARGET_DATASETandLABELwill be employed, to override them you can refer to the following commmand:make visualize SOURCE_DATASET=sample/real TARGET_DATASET=sample/fake LABEL=9
The main idea behind the working behaviour of the visualization is very simple since it extracts one random batch from both domains and looks for the images with the specified labels within it.
Therefore, there may be some cases in which the images with the wanted label are not contained and thus not shown, in this scenario I suggest you to increase the batch size or to run again the command.
We know that this functionality is limited and very naive but for our purposes it is more than enough.
To run the unsupervised domain adaptation experiment you can type:
python -m uda experiment POSITIONAL_ARGUMENTS [OPTIONAL_ARGUMENTS]whose arguments can be seen by typing:
python -m uda experiment --helpwhich are:
positional arguments:
{0,1,2,3,4,5,6,7,8,9}
which network to run, see the `Technique` enumerator in `uda/__init__.py` and select the one
you prefer
source_data source domain dataset
target_data target domain dataset
exp_name name of the experiment
num_classes number of classes [default 20]
pretrained pretrained model
epochs number of training epochs
{alexnet,resnet} backbone network
optional arguments:
-h, --help show this help message and exit
--wandb WANDB, -w WANDB
employ wandb to keep track of the runs
--device {cuda,cpu} device on which to run the experiment
--batch-size BATCH_SIZE
batch size
--test-batch-size TEST_BATCH_SIZE
test batch size
--learning-rate LEARNING_RATE
learning rate
--weight-decay WEIGHT_DECAY
sdg weight decay
--momentum MOMENTUM sdg momentum
--step-decay-weight STEP_DECAY_WEIGHT
sdg step weight decay
--active-domain-loss-step ACTIVE_DOMAIN_LOSS_STEP
active domain loss step
--lr-decay-step LR_DECAY_STEP
learning rate decay step
--alpha-weight ALPHA_WEIGHT
alpha weight factor for Domain Separation Networks
--beta-weight BETA_WEIGHT
beta weight factor for Domain Separation Networks
--gamma-weight GAMMA_WEIGHT
gamma weight factor for Domain Separation Networks
--save-every-epochs SAVE_EVERY_EPOCHS
how frequent to save the model
--reverse-domains REVERSE_DOMAINS
switch source and target domain
--dry DRY do not save checkpoints
--project-w PROJECT_W
wandb project
--entity-w ENTITY_W wandb entity
--classes CLASSES [CLASSES ...], -C CLASSES [CLASSES ...]
classes provided in the dataset, by default they are those employed for the projectwhere technique refers to:
class Technique(Enum):
r"""
Technique enumerator.
It is employed for letting understand which method to use
in order to train the neural network
"""
SOURCE_ONLY = 1 # Train using only the source domain
UPPER_BOUND = 2 # Train using the target domain too, to fix an upper bound
DDC = 3 # Deep Domain Confusion
DANN = 4 # Domain-Adversarial Neural Network
DSN = 5 # Domain Separation Network
ROTATION = 6 # Rotation Loss
MEDM = 7 # Entropy Minimization vs. Diversity Maximization
DANN_MEDM = 8 # DANN with Entropy Minimization vs. Diversity Maximization
DSN_MEDM = 9 # DSN with MEDMAs an effective representation the command:
python -m uda experiment 7 adaptiope_small/product_images adaptiope_small/real_life "trial" 20 pretrained 20 resnetwhich will produce something similar to the following result:
Alternatively, you can obtain the same result in a less verbose manner by tuning the flags in the Makefile and then run:
make experimentNote: The current behavior is equivalent to
python -m uda 7 adaptiope_small/product_images adaptiope_small/real_life "medm" 20 pretrained 20 resnet --learning-rate 0.001 --batch-size 32 --test-batch-size 32you can change the behaviour by passing the parameters to the
Makefileor by modifying its variables directly.
If you are interested in running all the experiments we have performed at once, we suggest you to execute the jupyter notebook in the repository.
Such notebook has been run on Google Colab, therefore you can effortless run it from there as a powerful enough GPU and all the required libraries are provided.
If you are willing to run the notebook be sure you have a copy of the zip archive of the Adaptiope dataset, which is available here, placed in your Google Drive folder inside a directory called datasets.
Practically speaking, the notebook will try to filter out the Adaptiope dataset by accessing My Drive/datasets/Adaptiope.zip after having established a connection to your Google Drive storage.