This repository contains the implementation of star-convex object detection for 2D and 3D images, as described in the papers:
-
Uwe Schmidt, Martin Weigert, Coleman Broaddus, and Gene Myers.
Cell Detection with Star-convex Polygons.
International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI), Granada, Spain, September 2018. -
Martin Weigert, Uwe Schmidt, Robert Haase, Ko Sugawara, and Gene Myers.
Star-convex Polyhedra for 3D Object Detection and Segmentation in Microscopy.
The IEEE Winter Conference on Applications of Computer Vision (WACV), Snowmass Village, Colorado, March 2020
Please cite the paper(s) if you are using this code in your research.
The following figure illustrates the general approach for 2D images. The training data consists of corresponding pairs of input (i.e. raw) images and fully annotated label images (i.e. every pixel is labeled with a unique object id or 0 for background). A model is trained to densely predict the distances (r) to the object boundary along a fixed set of rays and object probabilities (d), which together produce an overcomplete set of candidate polygons for a given input image. The final result is obtained via non-maximum suppression (NMS) of these candidates.
The approach for 3D volumes is similar to the one described for 2D, using pairs of input and fully annotated label volumes as training data.
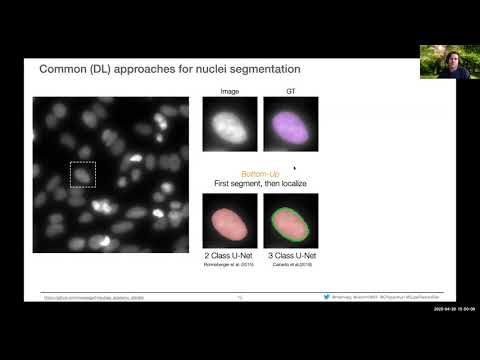
If you want to know more about the concepts and practical applications of StarDist, please have a look at the following webinar that was given at NEUBIAS Academy @Home 2020:
This package requires Python 3.6 (or newer).
-
Please first install TensorFlow (either TensorFlow 1 or 2) by following the official instructions. For GPU support, it is very important to install the specific versions of CUDA and cuDNN that are compatible with the respective version of TensorFlow.
-
StarDist can then be installed with
pip:pip install stardist
- Depending on your Python installation, you may need to use
pip3instead ofpip. - Since this package relies on a C++ extension, you could run into compilation problems (see Troubleshooting below). We currently do not provide pre-compiled binaries.
- StarDist uses the deep learning library Keras, which requires a suitable backend (we currently only support TensorFlow).
- (Optional) You need to install gputools if you want to use OpenCL-based computations on the GPU to speed up training.
- (Optional) You might experience improved performance during training if you additionally install the Multi-Label Anisotropic 3D Euclidean Distance Transform (MLAEDT-3D).
We provide example workflows for 2D and 3D via Jupyter notebooks that illustrate how this package can be used.
Currently we provide some pretrained models in 2D that might already be suitable for your images:
| key | Modality (Staining) | Image format | Example Image | Description |
|---|---|---|---|---|
2D_versatile_fluo 2D_paper_dsb2018 |
Fluorescence (nuclear marker) | 2D single channel |  |
Versatile (fluorescent nuclei) and DSB 2018 (from StarDist 2D paper) that were both trained on a subset of the DSB 2018 nuclei segmentation challenge dataset. |
2D_versatile_he |
Brightfield (H&E) | 2D RGB |  |
Versatile (H&E nuclei) that was trained on images from the MoNuSeg 2018 training data and the TNBC dataset from Naylor et al. (2018). |
You can access these pretrained models from stardist.models.StarDist2D
from stardist.models import StarDist2D
# prints a list of available models
StarDist2D.from_pretrained()
# creates a pretrained model
model = StarDist2D.from_pretrained('2D_versatile_fluo')To train a StarDist model you will need some ground-truth annotations: for every raw training image there has to be a corresponding label image where all pixels of a cell region are labeled with a distinct integer (and background pixels are labeled with 0). To create such annotations in 2D, there are several options, among them being Fiji, Labkit, or QuPath. In 3D, there are fewer options: Labkit and Paintera (the latter being very sophisticated but having a steeper learning curve).
Although each of these provide decent annotation tools, we currently recommend using Labkit (for 2D or 3D images) or QuPath (for 2D):
- Install Fiji and the Labkit plugin
- Open the (2D or 3D) image and start Labkit via
Plugins > Segmentation > Labkit - Successively add a new label and annotate a single cell instance with the brush tool (always check the
overrideoption) until all cells are labeled - Export the label image via
Save Labeling...andFile format > TIF Image
Additional tips:
- The Labkit viewer uses BigDataViewer and its keybindings (e.g. s for contrast options, CTRL+Shift+mouse-wheel for zoom-in/out etc.)
- For 3D images (XYZ) it is best to first convert it to a (XYT) timeseries (via
Re-Order Hyperstackand swappingzandt) and then use [ and ] in Labkit to walk through the slices.
- Install QuPath
- Create a new project (
File -> Project...-> Create project) and add your raw images - Annotate nuclei/objects
- Run this script to export the annotations (save the script and drag it on QuPath. Then execute it with
Run for project). The script will create aground_truthfolder within your QuPath project that includes both theimagesandmaskssubfolder that then can directly be used with StarDist.
To see how this could be done, have a look at the following example QuPath project (data courtesy of Romain Guiet, EPFL).
Installation requires Python 3.6 (or newer) and a working C++ compiler. We have only tested GCC (macOS, Linux), Clang (macOS), and Visual Studio (Windows 10). Please open an issue if you have problems that are not resolved by the information below.
If available, the C++ code will make use of OpenMP to exploit multiple CPU cores for substantially reduced runtime on modern CPUs. This can be important to prevent slow model training.
The default Apple C/C++ compiler (clang) does not come with OpenMP support and the package build will likely fail.
To properly build stardist you need to install an OpenMP-enabled GCC compiler, e.g. via Homebrew with brew install gcc (e.g. installing gcc-10/g++-10 or newer). After that, you can build the package like this (adjust compiler names/paths as necessary):
CC=gcc-10 CXX=g++-10 pip install stardist
If you use conda on macOS and after import stardist see errors similar to the following:
Symbol not found: _GOMP_loop_nonmonotonic_dynamic_next
please see this issue for a temporary workaround.
Please install the Build Tools for Visual Studio 2019 from Microsoft to compile extensions for Python 3.6 and newer (see this for further information). During installation, make sure to select the C++ build tools. Note that the compiler comes with OpenMP support.
We currently provide a ImageJ/Fiji plugin that can be used to run pretrained StarDist models on 2D or 2D+time images. Installation and usage instructions can be found at the plugin page.
@inproceedings{schmidt2018,
author = {Uwe Schmidt and Martin Weigert and Coleman Broaddus and Gene Myers},
title = {Cell Detection with Star-Convex Polygons},
booktitle = {Medical Image Computing and Computer Assisted Intervention - {MICCAI}
2018 - 21st International Conference, Granada, Spain, September 16-20, 2018, Proceedings, Part {II}},
pages = {265--273},
year = {2018},
doi = {10.1007/978-3-030-00934-2_30}
}
@inproceedings{weigert2020,
author = {Martin Weigert and Uwe Schmidt and Robert Haase and Ko Sugawara and Gene Myers},
title = {Star-convex Polyhedra for 3D Object Detection and Segmentation in Microscopy},
booktitle = {The IEEE Winter Conference on Applications of Computer Vision (WACV)},
month = {March},
year = {2020},
doi = {10.1109/WACV45572.2020.9093435}
}