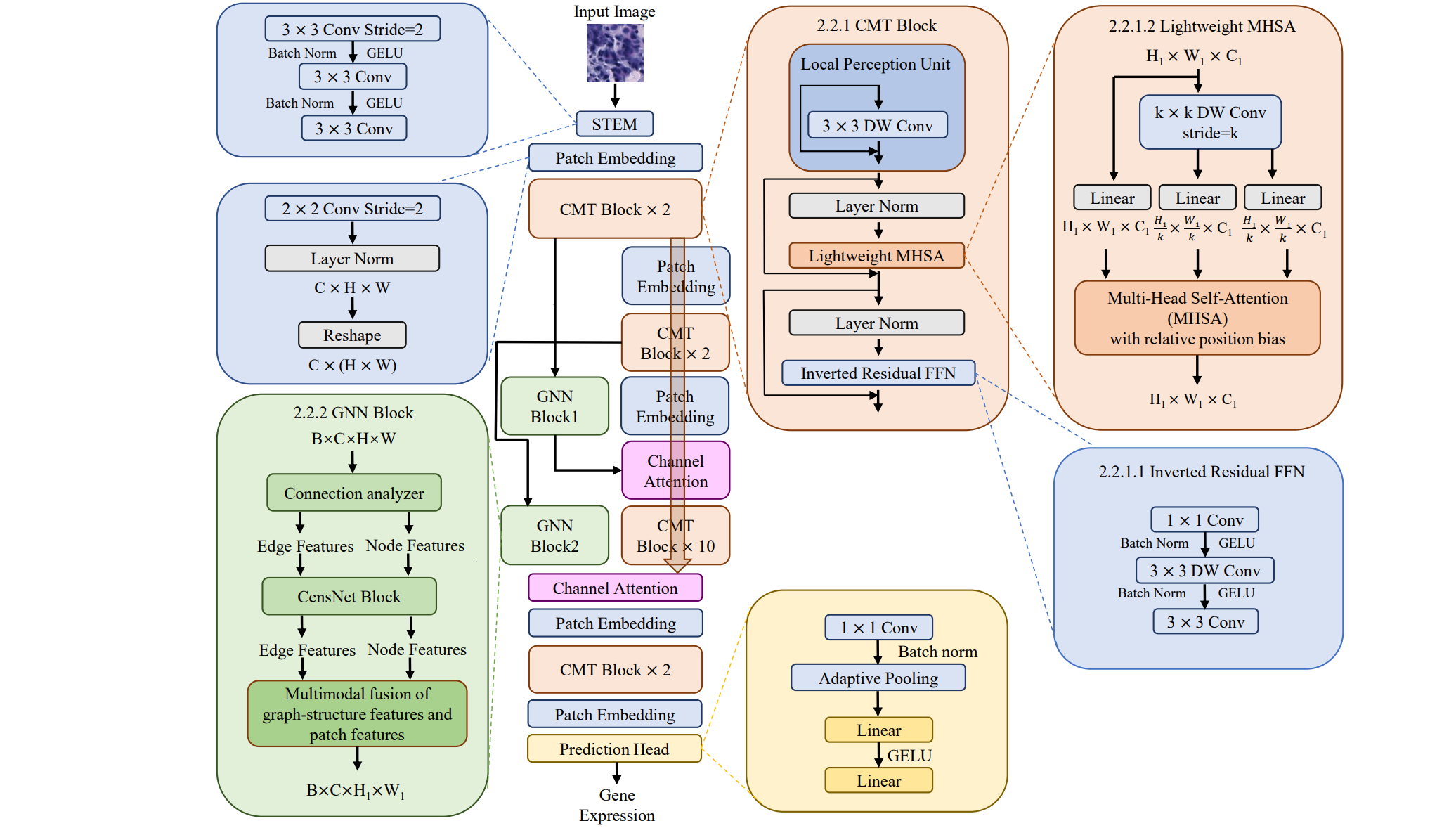
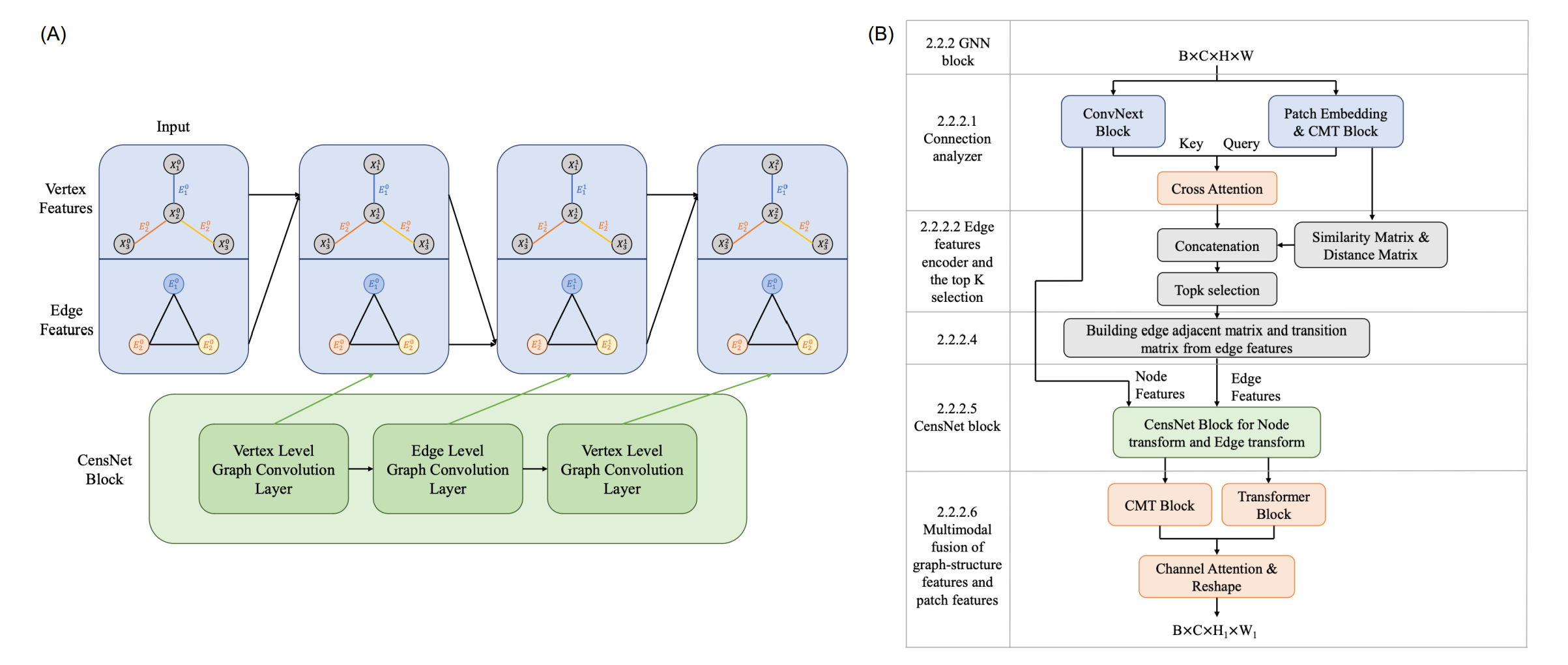
Transformer with Convolution and Graph-Node co-embedding: A accurate and interpretable vision backbone for predicting gene expressions from local histopathological image
Xiao Xiao1,2,3, Yan Kong1,2, Ronghan Li1,2, Zuoheng Wang3, Hui Lu2,1,4,*
1State Key Laboratory of Microbial Metabolism, Joint International Research Laboratory of Metabolic and Developmental Sciences, Department of Bioinformatics and Biostatistics, School of Life Sciences and Biotechnology, Shanghai Jiao Tong University, Shanghai, China
2SJTU-Yale Joint Center for Biostatistics and Data Science, National Center for Translational Medicine, Shanghai Jiao Tong University, Shanghai, China
3Department of Biostatistics, Yale University, New Haven, CT, United States
4Center for Biomedical Informatics, Shanghai Children’s Hospital, Shanghai, China
*** Correspondence:
** Hui Lu huilu@sjtu.edu.cn
Keywords: deep learning, breast cancer, convolutional neural network, graph neural network, transformer, spatial transcriptomics.
Highlights
-
SOTA gene expression prediction model using only one local image
-
Perform best on different human organs and can be applied to bulk RNA-seq
-
Generate important multimodal biomarker information from cheaper images
-
Accurately predicted genes are closely related to cancer immunology
-
First vision backbone to integrate CNN, GNN, and transformer for image analysis
As a general computer vision backbone, the TCGN model can be easily used.
import torch
import numpy as np
from model import TCGN
my_model=TCGN(num_classes=785)# predict 785 genes
# load pretrained model
my_model.load_state_dict(torch.load('./record-TCGN/A2-TCGN-her2-best.pth'), strict=True)
# use gpu to run the model
my_model = my_model.cuda()
# imgs: input batched image tensor on the gpu with shape Batch_size x 3 x 224 x 224
# if the magnification is 20x then orginal image size should be 112x112, and 56x56 for 10x magnification, then resize them to 224x224
# if the input img has the same size as the spot in the spatial transcriptomics and is not 224 x 224, then resize the img to 224 x 224
# img should be 0-1 normalized, then normalized by IMAGENET_DEFAULT_MEAN, IMAGENET_DEFAULT_STD in the python package timm
predictions=my_model(imgs).detach().cpu()
# predictions: predicted gene expression tensor with shape Batch_size x 785
# where 785 is the number of genes the model is set to output
# Specific for HER2+ dataset (breast cancer)
# Predict the expression of genes that can be statistically significantly predicted by the model
selected_genes_number_tensor=torch.Tensor(np.load("data/genes_her2_that_we_think_can_be_predicted.npy")).bool()
predictions=predictions[:,selected_genes_number_tensor]
# to numpy form if needed
predictions=predictions.numpy()Required package:
- PyTorch >= 1.10
- scanpy >= 1.8
- python >=3.7
- human HER2-positive breast tumor ST data https://github.com/almaan/her2st/.
- human cutaneous squamous cell carcinoma 10x Visium data: GSE144240 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE144240).
- SpatialLIBD dataset: http://research.libd.org/spatialLIBD/
- Please run the script
download.shin the folder data - Run
gunzip *.gzin the dir ``../data/her2st/data/ST-cnts/` to unzip the gz files - Run the following command to run the cross-validation on fold 0 (change the number 0 for other fold cross-validation):
python -u train.py --fold 0