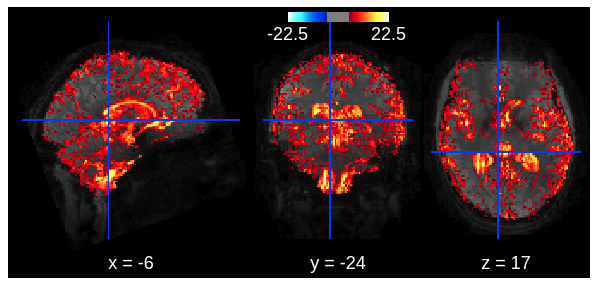
Code to convert Philips physiology files ("SCANPHYSLOG") to the BIDS-format, including the estimation of volume triggers using the logged gradient, volume markers, or by interpolation. It writes out BIDSified physio-files (as *.tsv.gz and associated *.json files). From there on, you can use other software to, for example, estimate RETROICOR/HRV/RVT regressors for nuisance regression. I recommend using the PhysIO toolbox for this (worked really well for me in the past, see image below), but FSL's PNM should also work.
I recommend installing the package from PyPI using pip:
pip install -U scanphyslog2bids
If you want the latest (development) version, you can install it from Github:
pip install https://github.com/lukassnoek/scanphyslog2bids/archive/master.zip
Or clone this repository locally and install as follows:
pip install .
This package uses Python 3.6 or higher and depends on the following Python packages:
- nibabel
- numpy
- pandas
- matplotlib (optional for plots)
- click (for the CLI)
This package comes with a Python interface and a CLI. See the code below for a minimal example using the Python interface:
from scanphyslog2bids.core import PhilipsPhysioLog
log_file = 'my_scanphyslog_file.log'
out_dir = '~/my_bids_data' # where the BIDSified data should be saved
deriv_dir '~/my_bids_data/physio' # where some QC plots should be saved
# fmri_file is used to extract metadata, such as TR and number of volumes
fmri_file = 'fmri_file_associated_with_scanphyslog.nii.gz'
fmri_img = nib.load(f)
n_dyns = fmri_img.shape[-1]
tr = np.round(fmri_img.header['pixdim'][4], 3)
# Create PhilipsPhysioLog object with info
phlog = PhilipsPhysioLog(f=log_file, tr=tr, n_dyns=ndyns, sf=496, manually_stopped=False)
# Load in data, do some preprocessing
phlog.load()
# Try to align physio data with scan data, using a particular method
# (either "vol_markers", "gradient_log", or "interpolation")
phlog.align(trigger_method='gradient_log') # load and find vol triggers
# Write out BIDS files
phlog.to_bids(out_dir) # writes out .tsv.gz and .json files
# Optional: plot some QC graphs for alignment and actual traces
phlog.plot_alignment(out_dir=deriv_dir) # plots alignment with gradient
phlog.plot_traces(out_dir=deriv_dir) # plots cardiac/resp tracesThe command line interface can be used as follows:
(base) lukas@uva:~/software/scanphyslog2bids$ scanphyslog2bids --help
Usage: scanphyslog2bids [OPTIONS]
Options:
--file TEXT Scanphyslog file to convert (mandatory)
--sf INTEGER Sampling rate (optional, default: 496)
--fmri TEXT Associated fmri file (optional, assuming ndyns and tr are given)
--ndyns INTEGER Number of dynamics/volumes (optional, assuming that fmri is given)
--tr FLOAT Repetition time of fmri scan (optional, assuming that fmri is given)
--manualstop Was the scan manually stopped? (optional, store True)
--triggermethod TEXT Method to detect triggers (optional, default: gradient_log)
--outdir TEXT Output directory for BIDS file (optional, default: parent-dir of phys-file)
--plottraces Whether to plot the traces (optional, store True)
--plotalignment Whether to plot the alignment (optional, store True)
--derivdir TEXT Derivatives directory (for plots) (optional, default:
--help Show this message and exit.
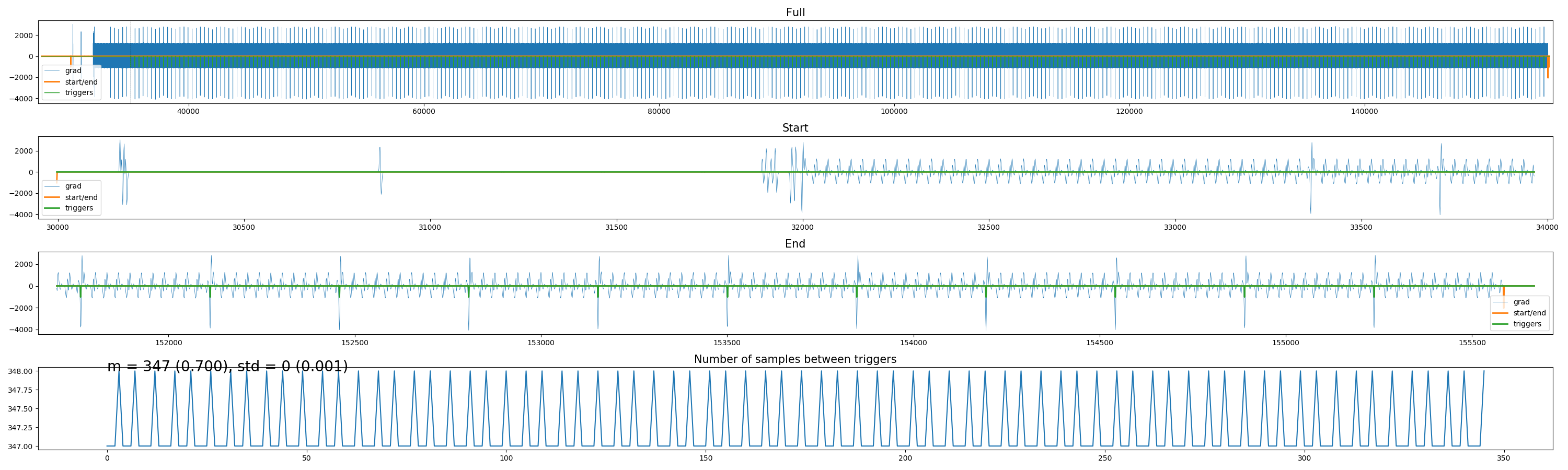
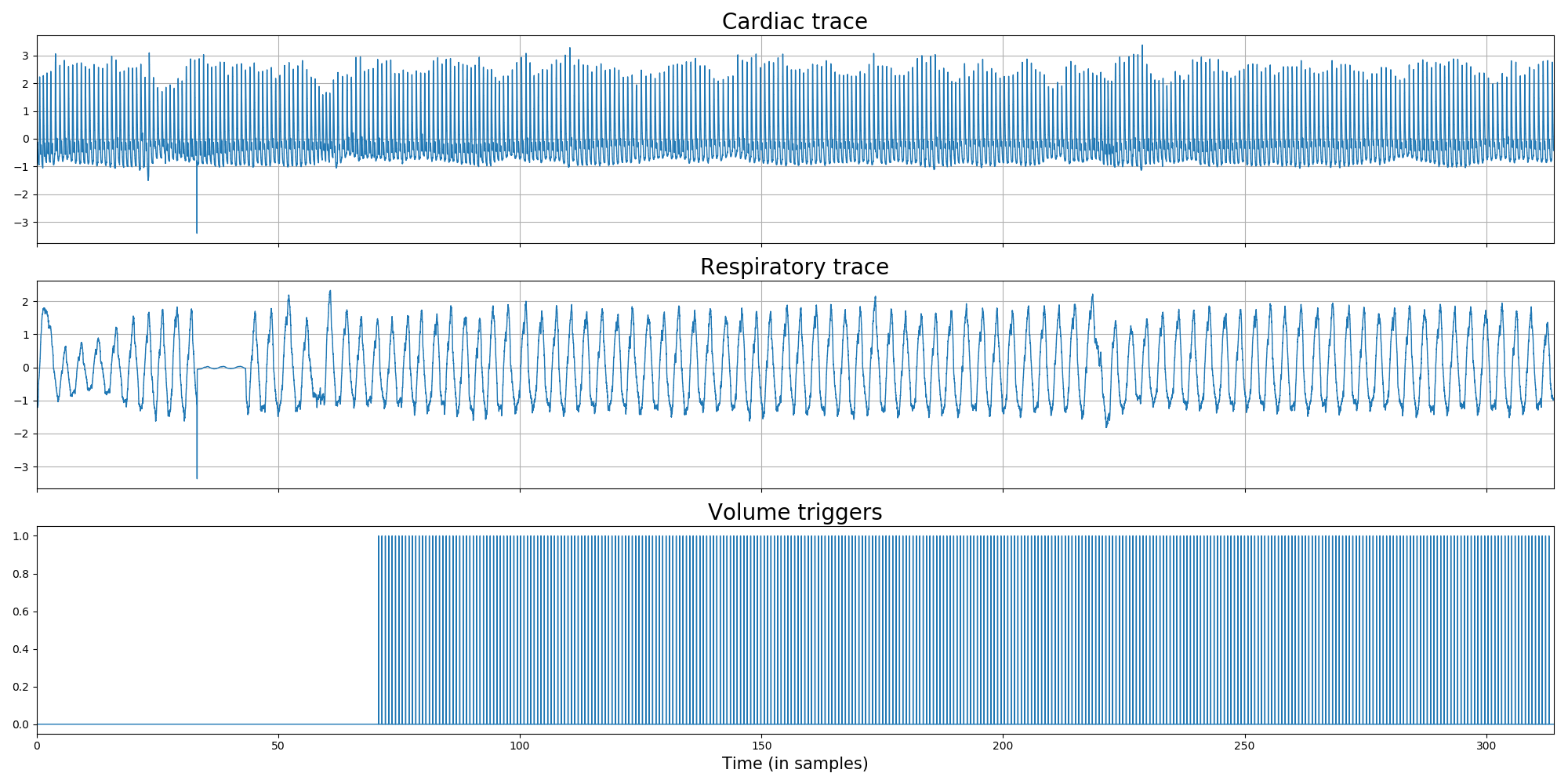
Apart from the BIDSified SCANPHYSLOG files (i.e., a *.tsv.gz with cardiac/respiratory/volume trigger traces and *.json file), the package allows for creating plots of the physio-scan alignment (using the plot_alignment method or the --plotalignment flag) and the actual respiratory and cardiac traces (using the plot_traces method or the --plottraces flag).
The alignment plot looks similar to the figure below, which visualizes the full gradient trace (if available) with the estimated volume triggers on top (first row), the close-up view of the start and end of the trace (second/third row; because here most issues tend to arise during alignment), and a trace of the number of samples in between (estimated) volume triggers (fourth row). The number of samples in between triggers should generally not deviate more than 2 samples across triggers.
You can also use this plot to detect funky gradients.
The "traces" plot allow you to do some visual QC on the respiratory and cardiac traces:
I recommend using the gradient_log method, which often works quite well, except for when your gradients are really funky (e.g., when you tilt the FOV a lot). For most (2D "ascending", i.e., inferior-superior or vice versa) scans, the "y" gradient direction works best to distill the volume onsets (use which_grad='y' when calling the align method).
If the gradients were not logged (this seems to happen at some scanners) or the gradient_log method fails, I'd recommend you use the interpolate trigger method. It works by "interpolating" triggers backwards from the end of the file (specifically from the "end marker"). This is, however, definitely not foolproof, as the "end marker" in SCANPHYSLOG files does not always seem to coincide with the actual offset of the last volume. I have found that this "offset" between the end of the last volume and the end marker may vary from 5 samples (~0.01 seconds) to about 166 samples (~0.332 seconds) depending on your scanner's hardware or software (interface) system. When using interpolation trigger method, you can control the assumed offset with the offset_end_scan parameter in the align method (for which the default is set to 20).
Also, make sure you use the right sampling frequency (the sf parameter). The default is set to 496 (Hz), which is the sampling frequency for wireless physio recorders. If you use a wired physio recorder, the sampling frequency is 500 Hz (see here).
To estimate RETROICOR (and HRV/RVT) regressors from the physiology data, I normally use the PhysIO toolbox. This toolbox should work with the BIDSified output from the scanphyslog2bids package. In the scripts/run_physIO.m file, I have included an example script which runs the PhysIO toolbox on an example output file from scanphyslog2bids. The settings there have worked for me in the past to succesfully create RETROICOR/HRV/RVT regressors for both 3T data (for 2D EPI scans) and 7T data (for 3D EPI scans).
Feel free to submit an issue when encountering issues (of better yet: send a PR if you fixed the bug yourself).