| Author: | Lukas Turcani |
|---|
Welcome to stk-vis!
Before you get started, you might like to watch the demo video
https://youtu.be/sIXu5W8o53Q
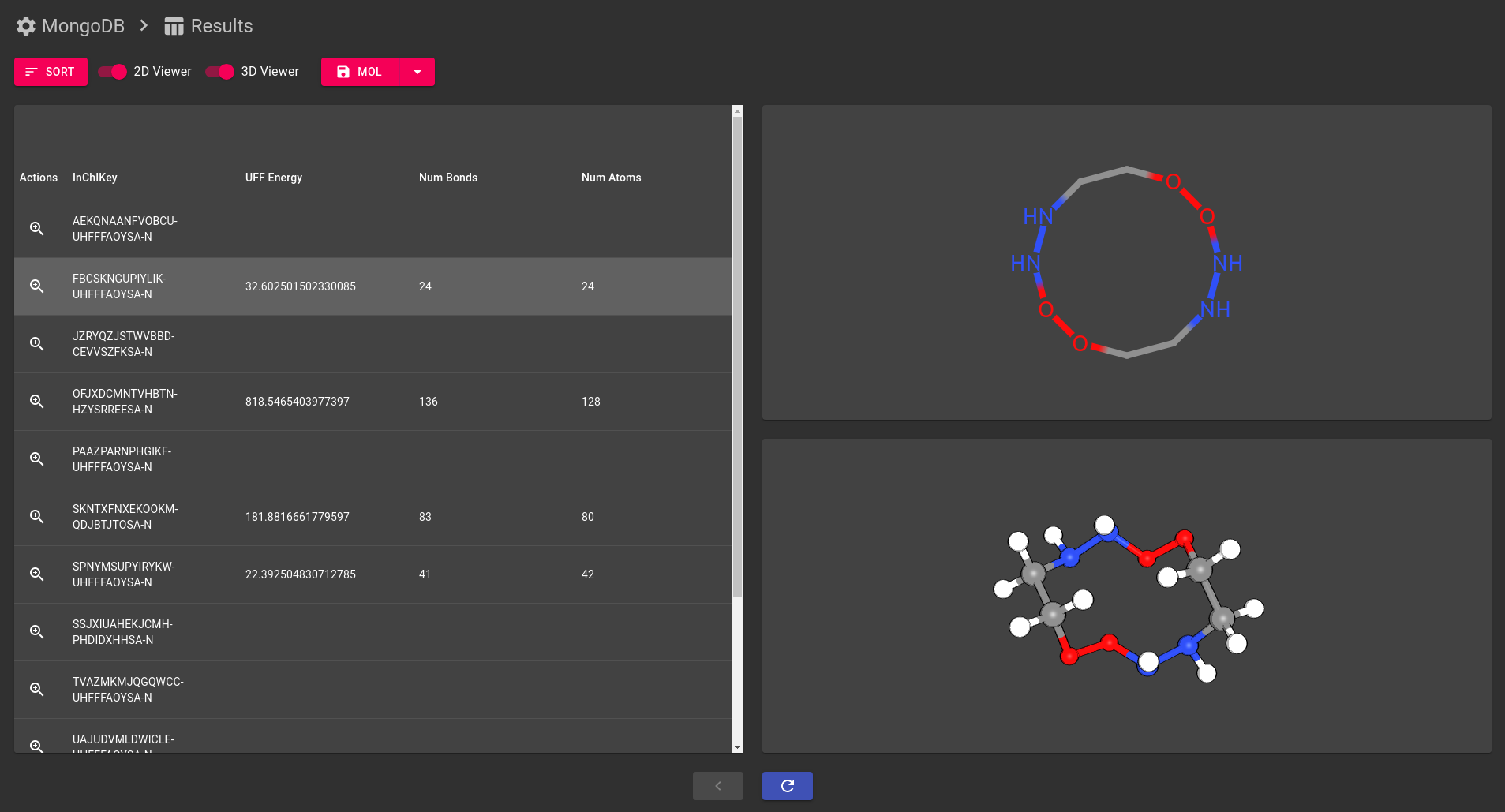
stk-vis is a cross-platform viewer for molecules and molecular
properties, specifically targeting cases where you want to browse
through multiple molecules quickly. Its main strength is allowing
smoother collaboration between multiple people or between multiple
research groups, but it is also a useful tool for individual users.
So how does stk-vis do this? stk-vis connects to local
or remote MongoDB molecular databases, which hold the molecules you
would like to view, as well as their properties. If you think
using a MongoDB database is a bit of a chore, I promise you that
it is actually trivial, that you don't need to know anything about
databases, and I will show you how to build one!
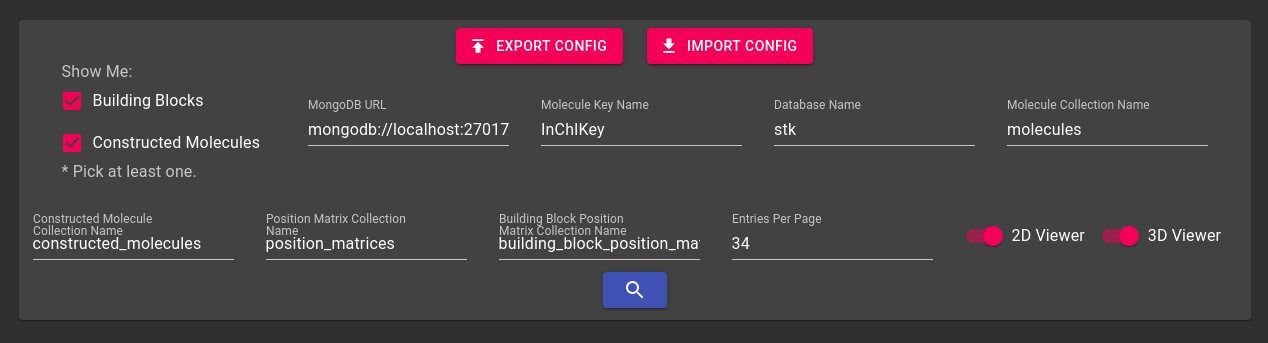
Assuming that you have your database, or someone has provided one
for you, you can connect to it from anywhere. For example, try
downloading the latest release of
stk-vis for your platform and using the following URL:
mongodb+srv://stk-vis:example@stk-vis-example.x4bkl.mongodb.net
Here is a picture of your connection settings:
This should connect you to a public database I made. Note that you don't have to change any of the other default values. (The connection might be a bit slow because I'm using a free server hosted in Europe, but when you use your own database it should be much faster.) You can also make your databases private and only allow access to specific users.
To give an example use case, you can have a group of computational
scientists depositing molecules and their properties into the database,
and their synthetic collaborators can immediately see which molecules
have been added and what their properties are, in order to see if they
would like to make anything. The synthetic group does not need
to worry about databases at all, they just need to download stk-vis
and connect to the URL the computational team provides them with.
Similarly, the computational team does not have worry too much about
databases either, as they can deposit molecules and their properties
into them with one very simple Python function.
In fact, any time you are dealing with lots of molecules, or lots of molecular files, it is probably a good idea to switch to using a database, whether you are working as an individual or not. This keeps all your data easy to access and organized.
- 3D interactive molecular rendering.
- 2D molecular projection.
- Tabulation of any molecular properties deposited into the database.
- If you have molecules that were constructed from building block molecules, you can see which building blocks were used to make that molecule, and then you can see if those building blocks had building blocks too! You can keep doing this until you run out of building blocks.
- Sort molecules according to property values to quickly find ones with the best or worst properties.
- Saving molecular structure files.
- Sometimes 2D projections are expensive to calculate. As a result, you can toggle the 2D viewer on or off. You can also toggle the 3D viewer, but I suggest you first check if the 2D viewer is the cause of performance problems.
You can see the latest release for your platform by clicking on the following link:
https://github.com/lukasturcani/stk-vis/releases
If you would like to get updates when a new version of stk-vis
is released, simply click on the watch button in the top right
corner of the GitHub page, and if you only care about new releases,
select Releases only from the dropdown menu.
So how do you actually get a molecular database? Well, there's a
Python library called stk, which lets you do just that. But not
only that, as stk lets you easily construct molecules such as
polymers, organic & metal-organic frameworks, organic and
metal-organic cages, metal complexes, rotaxanes and more!
The GitHub for stk is here
https://github.com/lukasturcani/stk
and the documentation can be found here
https://stk.readthedocs.io
stk lets you deposit regular or constructed molecules
into MongoDB molecular databases. stk molecules can also be
converted to and from rdkit molecules, if that's
convenient for your use-case. stk also provides evolutionary
algorithms (EAs) for molecular design. This means you can run the EA
and it will deposit the molecules it discovers into the database
for you, and you can use stk-vis to easily browse the results!
Assuming you have stk installed and MongoDB running locally,
building molecular databases is easy
import stk
import pymongo
import rdkit.Chem.AllChem as rdkit
client = pymongo.MongoClient()
db = stk.MoleculeMongoDb(
mongo_client=client,
# All of the parameters below are optional!
database='stk',
molecule_collection='molecules',
position_matrix_collection='building_block_position_matrices',
)
# Create some molecule. "BuildingBlock" is just stk's word for a
# plain molecule.
molecule1 = stk.BuildingBlock('CCCBr')
# Place it into the database, this will make the molecule
# immediately viewable in stk-vis.
db.put(molecule1)
# Make an stk molecule from an rdkit molecule and deposit it into
# the database. Note that the rdkit molecule must have a
# position matrix.
def get_rdkit_molecule(smiles):
molecule = rdkit.AddHs(rdkit.MolFromSmiles(smiles))
rdkit.EmbedMolecule(molecule, rdkit.ETKDGv2())
return molecule
molecule2 = stk.BuildingBlock.init_from_rdkit_mol(
molecule=get_rdkit_molecule('CNCNN'),
)
db.put(molecule2)stk provides detailed documentation for stk.MoleculeMongoDb.
Let's say you also want to deposit molecular properties into the
database so that they are available in stk-vis
num_atoms_db = stk.ValueMongoDb(client, 'Num Atoms')
# Place a value associated with the molecule into the database,
# this will make it immediately viewable in stk-vis.
num_atoms_db.put(molecule1, molecule1.get_num_atoms())
num_atoms_db.put(molecule2, molecule2.get_num_atoms())
# Lets also calculate and store the energy of a molecule with
# UFF.
def uff_energy(molecule):
rdkit_molecule = molecule.to_rdkit_mol()
rdkit.SanitizeMol(rdkit_molecule)
ff = rdkit.UFFGetMoleculeForceField(rdkit_molecule)
return ff.CalcEnergy()
energy_db = stk.ValueMongoDb(client, 'UFF Energy')
energy_db.put(molecule1, uff_energy(molecule1))
energy_db.put(molecule2, uff_energy(molecule2))In general, you can deposit any number or string, or
tuple of them
into a stk.ValueMongoDb. stk also has detailed documentation
for stk.ValueMongoDb
Finally, let's take a look at depositing constructed molecules.
These are molecules stk can construct from BuildingBlock
molecules. There are many different kinds of these molecules, so
check out the documentation of stk to get a full picture.
However, when it comes to depositing them into a MongoDB, the process
is always the same.
# Create a database for depositing constructed molecules.
constructed_db = stk.ConstructedMoleculeMongoDb(
mongo_client=client,
# All of the parameters below are optional!
database='stk',
molecule_collection='molecules',
constructed_molecule_collection='constructed_molecules',
position_matrix_collection='position_matrices',
building_block_position_matrix_collection='building_block_position_matrices',
)
# Create a constructed molecule, in this case a polymer.
polymer = stk.ConstructedMolecule(
topology_graph=stk.polymer.Linear(
building_blocks=(
stk.BuildingBlock('BrC=CBr', [stk.BromoFactory()]),
stk.BuildingBlock('BrCNCBr', [stk.BromoFactory()]),
),
repeating_unit='AB',
num_repeating_units=2,
),
)
# Deposit into the database.
constructed_db.put(polymer)
# You can deposit values same as before.
num_atoms_db.put(polymer, polymer.get_num_atoms())
energy_db.put(polymer, uff_energy(polymer))The reason stk.ConstructedMoleculeMongoDb is used here, is that
it will automatically deposit the building blocks of polymer into
the database as well. This means that in stk-vis, we can explicitly
search for the building blocks of polymer. As before, stk has
detailed documentation for stk.ConstructedMoleculeMongoDb.
To get stk you need to run:
$ pip install stk $ conda install -c conda-forge rdkit
Finally, you need to decide how to host your databases. You can
install MongoDB locally on your computer, or you can use
Mongo Atlas to put your database in the cloud. This part might be a
pain, but it shouldn't be too difficult either. Once this is
done, depositing molecules and molecular properties into the database
will be super easy with stk, and then you and your collaborators
can then examine them with stk-vis!