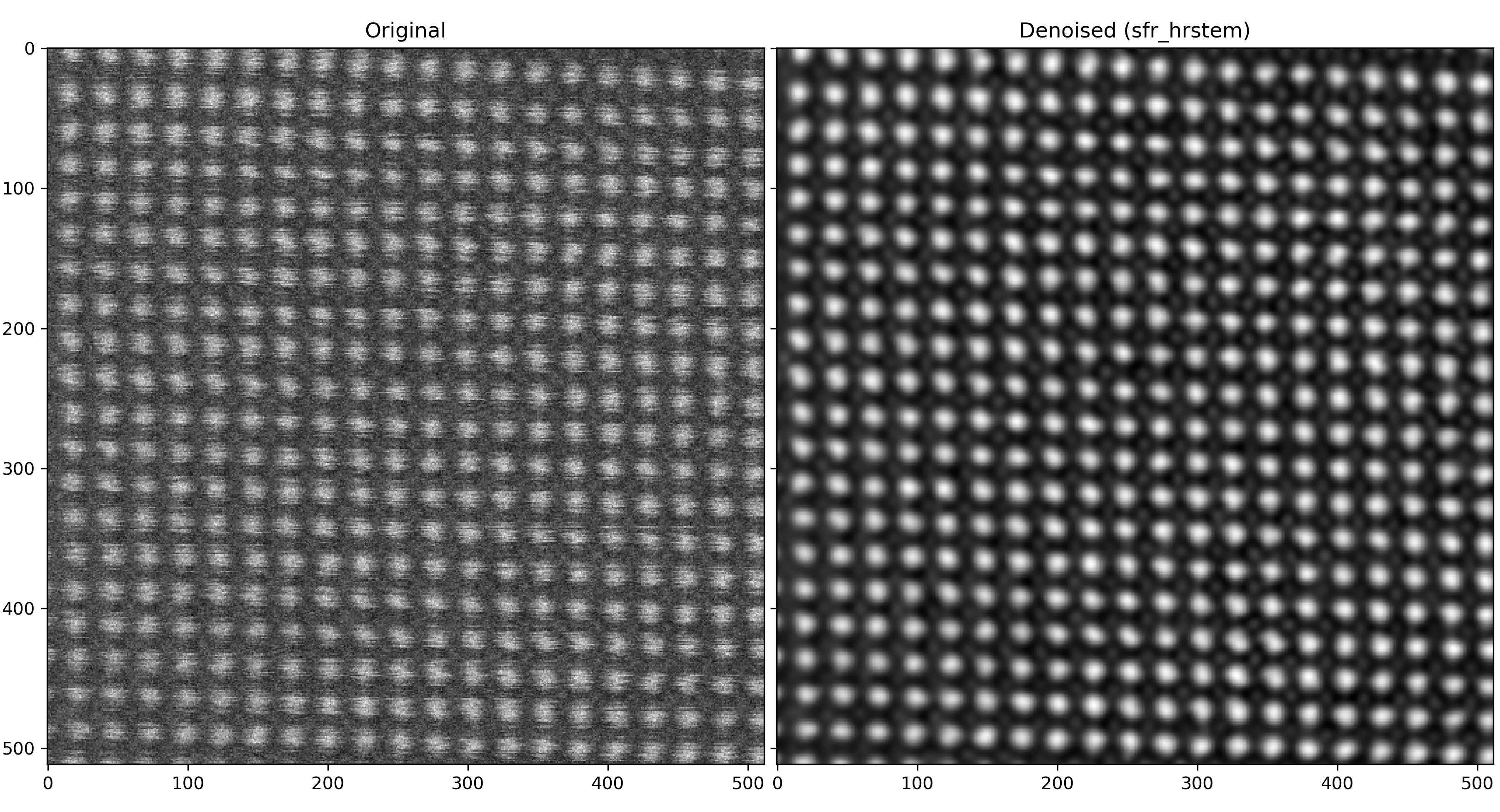
This notebook contains ideas for denoising workflows using the model and guidelines from the official repository (https://github.com/Ivanlh20/r_em).
It shows how to use tifffile to load TIFF images, denoise/restore them, and save them back to TIFF.
There is also an example showing how to use RosettaSciIO to directly read .emi/.ser or .dm3/.dm4 files to numpy array for subsequent denoising.
Paper: https://www.nature.com/articles/s41524-023-01188-0
Please cite their work in your publications if it helps your research:
@article{Lobato2024,
author = {I. Lobato and T. Friedrich and S. Van Aert},
doi = {10.1038/s41524-023-01188-0},
issn = {2057-3960},
issue = {1},
journal = {npj Computational Materials 2024 10:1},
keywords = {Imaging techniques,Transmission electron microscopy},
month = {1},
pages = {1-19},
publisher = {Nature Publishing Group},
title = {Deep convolutional neural networks to restore single-shot electron microscopy images},
volume = {10},
url = {https://www.nature.com/articles/s41524-023-01188-0},
year = {2024},
}Example of SrTiO$_3$[100] (HAADF, FEI Tecnai Osiris, 200 keV):