THIS IS AN EARLY AND BUGGY ALPHA RELEASE NOT INTENDED FOR DISTRIBUTION!
- Number of jobs in cluster submission may not be smaller then number of scaffolds in the assembly
- Neither number of jobs nor number scaffolds may be one
- Needs to be able to parse various lanes/readfiles in same protocol
- Need to organize all output of each stage in folders (it's rather messy right now)
- Organize module structure and separate from driver
- Need's a lot more input validation
- A deamon to automate submission of scattered stages would be nice
- do a dedup stage in first pass and extract reads from that for the follwing one?
Raccoon is a semi automated pipeline that is intended to be deployed as the last step of a reference genome assembly in order to correct base substitutions and small indels ('polishing'). It takes as input the raw data used to assemble the reference, as well as the assembly itself. The reads are used to perform variant calls of single nucleotide variants and short (<7bp) indels. After hard filtering, the called variants are integrated into the reference and the raw data is remapped onto the modifed assembly. Decreases in observed read depths of full length matching reads in regions were a variant has been integrated are used as a criterium to reject wronlgy performed error corrections. This process is performed iteratively. Polishing reference assemblies is currently particularly important for regions where PacBio data has been used, as these contain an elevated amount of indels even after self correction with quiver.
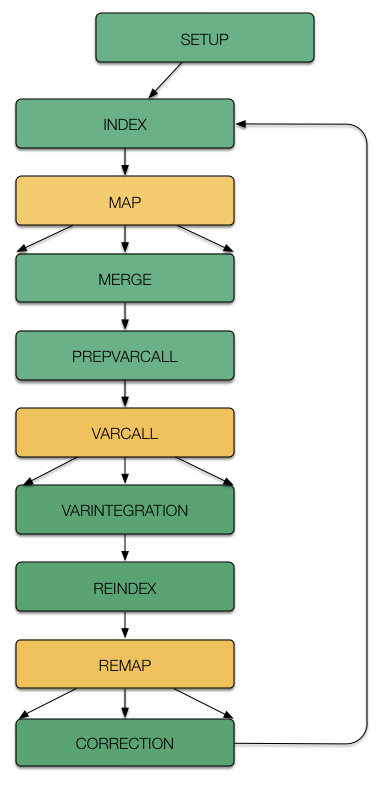
Raccoon is implemeted as a driver with accessory scripts, that executes or automatically submits to a distributed cluster. One pass of error corrections is comprised of 12 stages, with one additionall setup stage that only needs to be executed once. Wherever possible, the stages have been parallelized via a scatter-gather approach to make error corrections of large genomes feasible. Each stage automatically executes the follwing one until it hits a scattered stage. These scattered stages are highlited in yellow in the follwing schmee. The user only needs to execute stages follwing a scattered stage.
The basic algorithm upon which raccoon builds has first been described by Otto et al., 2010. However, its current implementation (iCORN2) fails to scale for genomes beyond 300-400Mb, a limitation that we seek to overcome with raccoon. Raccoon is implemented completely from scrach and does not build on iCorns' codebase.
Racoon is written in python 3 (tested with version 3.4). It currently depends on the follwing external programs (parenthesis denote the version used in development. This are the versions you should use, any other ones are not guaranteed to work):
Furthermore, the follwing non-default python3 modules are required:
For the actual pipeline, there is currently no installation process. Just download the folder and export it to you $PATH like so:
cd /path/to/raccoon
export PATH=$PATH:$(pwd)
For now, it is essential that the raccoon program remains within this folder, as its modules can otherwise not be accessed.
The pipeline is run through the driver like so:
raccoon stage protocol
The stage argument is one of the follwing 12:
- setup
- index
- map
- merge
- prepvarcall
- varcall
- varintegration
- reindex
- remap
- remerge
- correction
The setup stage only needs to be run once at the very beginning. The remaining stages can be run iteratively, and raccoon will automatically take care of rewiring the input for each iteration. By default, each stage (except setup) will automatically call the following stages once it finishes, until a scattered stage is reached (denoted in bold letters above). This means that calling index will automatically call index and map. Calling map will call call merge, prepvarcall and varcall. Calling varintegration will call varintegration, reindex and remap. Calling remerge will call remerge, correction and subsequently index and map for the following iteration. If you want to call each stage manually for some reasone (e.g. an intermediate stage failed), this can be done by invoking the -p argument, like so:
raccoon stage protocol -p
The protocol file is an xml file setting various paramteres and paths. The following shows a template protocol and explains the individual tags. All uppercase content are integers
<ec_pipeline>
<reference>/path/to/reference.fa</reference>
<outputDir>/path/to/output/basedir/</outputDir>
<input baseDir='/path/to/input/basedir'>
<p1>readFile.pair1.fastq</p1>i
<p2>readFile.pair2.fastq</p2>
<nPairs>NUMBER OF READS/READ-PAIRS</nPairs>
</input>
<ploidy>PLOIDY</ploidy>
<threads>NUMBER OF THREADS</threads>
<cluster>
<nJobs>NUMBER OF JOBS</nJobs>
<template>your-cluster-scheduler -n ${JOBNAME} -standard_error ${STDERR} -standard_output ${STDOUT} -command "${CMD}"</template>
</cluster>
<paths>
<scripts>/racoon/basedir</scripts>
<bwa>/path/to/bwa</bwa>
<samtools>/path/to/samtools</samtools>
<picardtools>/path/to/picard.jar</picardtools>
<gatk>/path/to/GenomeAnalysisTK.jar</gatk>
<python3>/path/to/python3</python3>
<java>/path/to/java</java>
</paths>
</ec_pipeline>
The <ec_pipeline> tags delimit the protocol.
The <reference> tag points to the reference assembly to be corrected.
The <outputDir> tag points to the base directory where the output will be stored.
The <input baseDir='x'> points to the base directory (x) were your read files are stored. It contains further nested tags: <p1> and <p2> point to the actual readfiles. If <p2> is ommited, the input is treated as single end. The <nPairs> contains the number of read pairs (or reads, if run with single end data). This information is necessary to know how big the chunks for scattered stages will be.
The <ploidy> tag denotes the ploidy of your genome.
In order to take care of automatic cluster submission, raccoon needs to know to submit to you cluster. This is done via the <cluster> tag. It contains two nested tags: <nJobs> denotes how many parallel jobs should be submitted. <template>gives a template string for cluster submission, in which the variable ${JOBNAME}, ${STDERR}, ${STDOUT}, ${CMD} will be replaced with the relevant values. The user must provide the template string around these values. For example: if your cluster is running torque, the content of your template tag should look like this:
<template>qsub -N ${JOBNAME} -e ${STDERR} -o ${STDOUT} -ADDITIONAL PARAMETERS ${CMD}</template>
The specific string depends on your system.
raccoon depends on several programs internally. While most of them are likely to be standard isntallations on systems within genomic research institutes, their default version migth not be the required ones. In order to overcome this problem easily, the direct path to the executable, jar or folder can be provided in the protocol. The valid tags are <scripts> (to point to raccoons base directory), <bwa>, <samtools>, <picardtools>, <gatk>, <python3> and <java>.
I didn't write this section yet, because it will change as soon as i tidy some things up. The important part for now is: A basedirectory will be created. It will contain a folder called READS containing the output of the setup stage (chunked up readfiles for parallelisation) as well as one folder for each iteration raccoon performs. The error corrected assembly is called reference.sanitizedVariants.fa