The dataset can be accessed via this link. This dataset can only be used for non-commercial research purposes. At no time, the dataset shall be used for clinical decisions or patient care. The data use license is CC BY-NC-ND 4.0.
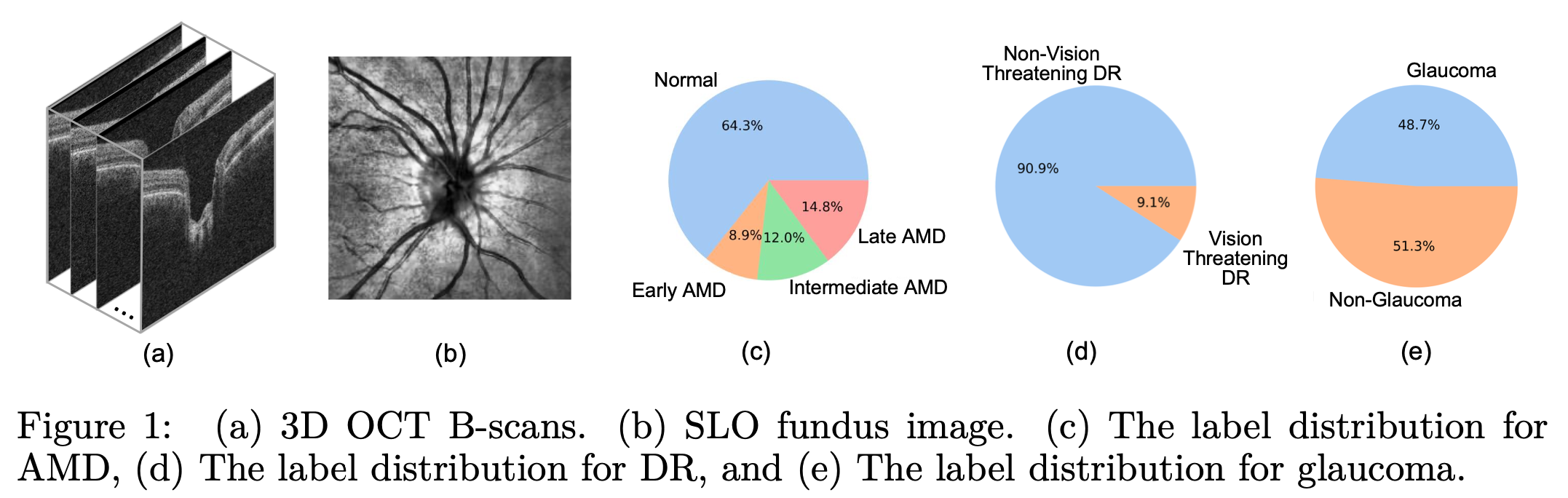
Our dataset includes 10,000 subjects for Age-Related Macular Degeneration (AMD), Diabetic Retinopathy (DR), and glaucoma separately, totaling 30,000 subjects with comprehensive demographic identity attributes including age, gender, race, ethnicity, preferred language, and marital status. Each subject has one Scanning Laser Ophthalmoscopy (SLO) fundus photo and one sample of Optical Coherence Tomography (OCT) B-scans. The size of OCT B-scans is 200 x 200 x 200 in glaucoma, while the one of OCT B-scans. The size of OCT B-scans is 128 x 200 x 200 in AMD and DR.
The dataset has an approximate size of 600 GB. Upon downloading and extracting these datasets, the names of the folders in the downloaded dataset under folders AMD/DR/Glaucoma are Training, Validation, and Test, respectively, for readability. Please rename them as train, val, and test. Then, you will find the dataset structure as follows.
FairVision
├── AMD
│ ├── test
│ ├── train
│ └── val
├── data_summary_amd.csv
├── DR
│ ├── test
│ ├── train
│ └── val
├── data_summary_dr.csv
├── Glaucoma
│ ├── test
│ ├── train
│ └── val
└── data_summary_glaucoma.csv
The "train/val/test" directories contain two types of data: SLO fundus photos and NPZ files that store OCT B-scans, SLO fundus photos, and additional attributes. SLO fundus photos serve visual inspection purposes, while the copies in NPZ files eliminate the need for the dataloader to access any other files except the NPZ files. The naming convention for SLO fundus photos follows the format "slo_xxxxxx.jpg," and for NPZ files, it is "data_xxxxx.npz," where "xxxxx" (e.g., 06691) represents a unique numeric ID. The dimensions of SLO fundus photos in NPZ files are 200 x 200, whereas those in the train/val/test folders are 512 x 664. The SLO fundus photos in NPZ files are created by resizing the photos in the folders and then normalizing them to [0, 255].
NPZ files have the following keys.
In the AMD disease, the NPZ files have
dr_subtype: AMD conditions - {'not.in.icd.table', 'no.amd.diagnosis', 'early.dry', 'intermediate.dry', 'advanced.atrophic.dry.with.subfoveal.involvement', 'advanced.atrophic.dry.without.subfoveal.involvement', 'wet.amd.active.choroidal.neovascularization', 'wet.amd.inactive.choroidal.neovascularization', 'wet.amd.inactive.scar'}
oct_bscans: images of OCT B-scans
slo_fundus: image of SLO fundus
race: 0 - Asian, 1 - Black, 2 - White
male: 0 - Female, 1 - Male
hispanic: 0 - Non-Hispanic, 1 - Hispanic
maritalstatus: 0 - Married, 1 - Single, 2 - Divorced, 3 - Widowed, 4 - Leg-Sep
language: 0 - English, 1 - Spanish, 2 - Others
The condition would be converted into the label of AMD by the condition-disease mapping.
condition_disease_mapping = {'not.in.icd.table': 0.,
'no.amd.diagnosis': 0.,
'early.dry': 1.,
'intermediate.dry': 2.,
'advanced.atrophic.dry.with.subfoveal.involvement': 3.,
'advanced.atrophic.dry.without.subfoveal.involvement': 3.,
'wet.amd.active.choroidal.neovascularization': 3.,
'wet.amd.inactive.choroidal.neovascularization': 3.,
'wet.amd.inactive.scar': 3.}
In the DR disease, the NPZ files have
dr_subtype: DR conditions - {'not.in.icd.table', 'no.dr.diagnosis', 'mild.npdr', 'moderate.npdr', 'severe.npdr', 'pdr'}
oct_bscans: images of OCT B-scans
slo_fundus: image of SLO fundus
race: 0 - Asian, 1 - Black, 2 - White
male: 0 - Female, 1 - Male
hispanic: 0 - Non-Hispanic, 1 - Hispanic
maritalstatus: 0 - Married, 1 - Single, 2 - Divorced, 3 - Widowed, 4 - Leg-Sep
language: 0 - English, 1 - Spanish, 2 - Others
The condition would be converted into the label of vision-threatening DR by the condition-disease mapping.
condition_disease_mapping = {'not.in.icd.table': 0.,
'no.dr.diagnosis': 0.,
'mild.npdr': 0.,
'moderate.npdr': 0.,
'severe.npdr': 1.,
'pdr': 1.}
In the glaucoma disease, the NPZ files have
glaucoma: the label of glaucoma disease, 0 - non-glaucoma, 1 - glaucoma
oct_bscans: images of OCT B-scans
slo_fundus: image of SLO fundus
race: 0 - Asian, 1 - Black, 2 - White
male: 0 - Female, 1 - Male
hispanic: 0 - Non-Hispanic, 1 - Hispanic
maritalstatus: 0 - Married, 1 - Single, 2 - Divorced, 3 - Widowed, 4 - Leg-Sep
language: 0 - English, 1 - Spanish, 2 - Others
We put all the attributes associated with the 10,000 samples in a meta csv file for each disease, including race, gender, ethnicity, marital status, age, preferred language.
Equity in AI for healthcare is crucial due to its direct impact on human well-being. Despite advancements in 2D medical imaging fairness, the fairness of 3D models remains underexplored, hindered by the small sizes of 3D fairness datasets. Since 3D imaging surpasses 2D imaging in SOTA clinical care, it is critical to understand the fairness of these 3D models. To address this research gap, we conduct the first comprehensive study on the fairness of 3D medical imaging models across multiple protected attributes. Our investigation spans both 2D and 3D models and evaluates fairness across five architectures on three common eye diseases, revealing significant biases across race, gender, and ethnicity. To alleviate these biases, we propose a novel fair identity scaling (FIS) method that improves both overall performance and fairness, outperforming various SOTA fairness methods. Moreover, we release Harvard-FairVision, the first large-scale medical fairness dataset with 30,000 subjects featuring both 2D and 3D imaging data and six demographic identity attributes. Harvard-FairVision provides labels for three major eye disorders affecting about 380 million people worldwide, serving as a valuable resource for both 2D and 3D fairness learning.
To install the prerequisites, run:
pip install -r requirements.txt
To run the experiments with the baseline models (e.g., ViT) on the task of AMD detection with SLO fundus images, execute:
./scripts/train_amd_vit.sh
To run the experiments with the baseline models (e.g., ViT) with the proposed FIS on the task of AMD detection with SLO fundus images, execute:
./scripts/train_amd_vit_fis.sh
To run the experiments with 3D ResNet on the task of AMD detection with OCT B-Scans, execute:
./scripts/train_amd_3d.sh
To run the experiments with 3D ResNet with the proposed FIS on the task of AMD detection with OCT B-Scans, execute:
./scripts/train_amd_3d_fis.sh
To run the experiments on the tasks of DR and Glaucoma detection, you can edit the aforementioned scripts by changing the dataset directory (e.g., FairVision/AMD -> FairVision/DR) and the python file (e.g., train_amd_fair.py/train_amd_fair_fis.py/train_amd_fair_3d.py/train_amd_fair_3d_fis.py -> train_dr_fair.py/train_dr_fair_fis.py/train_dr_fair_3d.py/train_dr_fair_3d_fis.py).