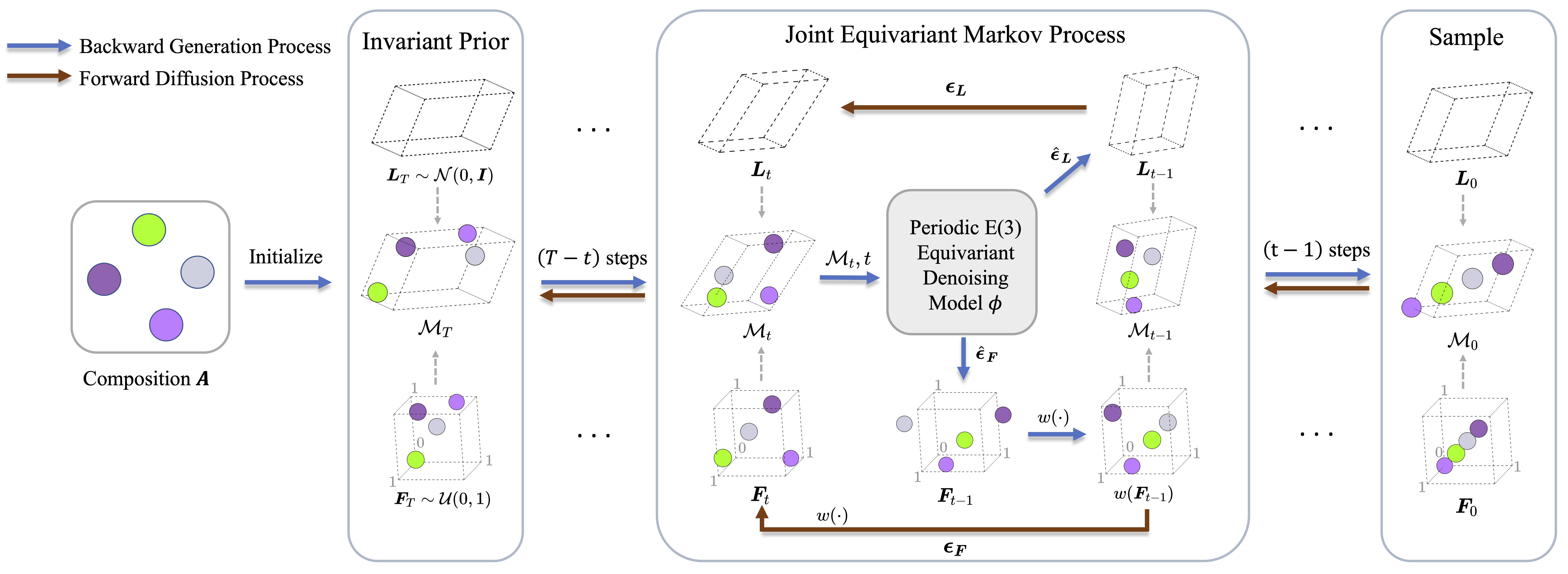
Implementation codes for Crystal Structure Prediction by Joint Equivariant Diffusion (DiffCSP).
Note: updated python==3.11 and pytorch>=2.0.0 and hydra>=1.3
# sudo apt-get install gfortran libfftw3-dev pkg-config
conda create -n diffcsp python=3.11.9
conda activate diffcsp
pip install torch==2.3.1 torchvision torchaudio --index-url https://download.pytorch.org/whl/cu121
pip install torch_geometric==2.5.3
pip install pyg_lib torch_scatter torch_sparse torch_cluster torch_spline_conv -f https://data.pyg.org/whl/torch-2.3.0+cu121.html
pip install lightning==2.3.2
pip install hydra-core omegaconf python-dotenv wandb rich
pip install p_tqdm pymatgen pyxtal smact matminer einops chemparse torchdyn
pip install -e .
mkdir logRename the .env.template file into .env and specify the following variables.
PROJECT_ROOT: the absolute path of this repo
HYDRA_JOBS: the absolute path to save hydra outputs
WANDB_DIR: the absolute path to save wabdb outputs
example:
# vi .env
export PROJECT_ROOT="/home/<YOURHOME>/DiffCSP"
export HYDRA_JOBS="/home/<YOURHOME>/DiffCSP/hydra"
export WANDB_DIR="/home/<YOURHOME>/DiffCSP/log"For the CSP task
python diffcsp/run.py data=<dataset> expname=<expname>
# example:
# python diffcsp/run.py data=mp_20 logging.wandb.group=mp_20 expname=originFor the Ab Initio Generation task
python diffcsp/run.py data=<dataset> model=diffusion_w_type expname=<expname>The <dataset> tag can be selected from perov_5, mp_20, mpts_52 and carbon_24, and the <expname> tag can be an arbitrary name to identify each experiment. Pre-trained checkpoints are provided here.
optional command line parameters
model=[flow|flow_polar] # use direct lattice or polar-decomposition lattice
+model.from_cubic=true # sample lattice from cubic prior, only effect on flow_polar
+lattice_polar_sigma=1 # lattice polar prior sigma
model.decoder.rec_emb=sin model.decoder.num_millers=6 # use reciprocal representation
model.time_dim=0 # if 0, use no embedding, else sinusoidal embedding
+model.lattice_teacher_forcing=-1 # epoch of teacher-forcing on lattice
model.cost_coord=10 model.cost_lattice=0.1 # fix coords or lattice if less than 1e-5
+model.ot=true # use optimal transport, default false
example for ab-init model training
model=flow_polar_w_type # w_type
model.cost_type=100 model.cost_coord=100 model.cost_lattice=1
One sample
python scripts/evaluate.py --model_path <model_path> --dataset <dataset>
python scripts/compute_metrics.py --root_path <model_path> --tasks csp --gt_file data/<dataset>/test.csv
# example:
# python ~/DiffCSP/scripts/evaluate.py --model_path `pwd` --dataset mp_20
# python ~/DiffCSP/scripts/compute_metrics.py --root_path `pwd` --tasks csp --gt_file ~/DiffCSP/data/mp_20/test.csvMultiple samples
python scripts/evaluate.py --model_path <model_path> --dataset <dataset> --num_evals 20
python scripts/compute_metrics.py --root_path <model_path> --tasks csp --gt_file data/<dataset>/test.csv --multi_evalpython scripts/generation.py --model_path <model_path> --dataset <dataset>
python scripts/compute_metrics.py --root_path <model_path> --tasks gen --gt_file data/<dataset>/test.csvpython scripts/sample.py --model_path <model_path> --save_path <save_path> --formula <formula> --num_evals <num_evals># train a time-dependent energy prediction model
python diffcsp/run.py data=<dataset> model=energy expname=<expname> data.datamodule.batch_size.test=100
# Optimization
python scripts/optimization.py --model_path <energy_model_path> --uncond_path <model_path>
# Evaluation
python scripts/compute_metrics.py --root_path <energy_model_path> --tasks opt
for solver in euler midpoint rk4; do
for seq in lf cf; do
for N in 20 50; do
label=N${N}R1S${solver}-${seq}
echo $label
CUDA_VISIBLE_DEVICES=0 python ~/DiffCSP/scripts/evaluate_ode.py \
--model_path `pwd` -N $N --solver $solver -seq $seq \
--test_bs 1000 --label $label
# legacy metrics
python ~/DiffCSP/scripts/compute_metrics.py --root_path `pwd` --label $label
# metrics without SMACT
# python ~/DiffCSP/scripts/compute_metrics_rec.py --root_path `pwd` --label $label
done
done
doneCUDA_VISIBLE_DEVICES=0 python ~/DiffCSP/scripts/sample_ode.py -m `pwd` -d sample -hpython scripts/generation.py --model_path <model_path> --dataset <dataset> --step_lr 0.01
python scripts/compute_metrics.py --root_path <model_path> --tasks gen --gt_file data/<dataset>/test.csvThe main framework of this codebase is build upon CDVAE. For the datasets, Perov-5, Carbon-24 and MP-20 are from CDVAE, and MPTS-52 is collected from its original codebase.
Please consider citing our work if you find it helpful:
@article{jiao2023crystal,
title={Crystal structure prediction by joint equivariant diffusion},
author={Jiao, Rui and Huang, Wenbing and Lin, Peijia and Han, Jiaqi and Chen, Pin and Lu, Yutong and Liu, Yang},
journal={arXiv preprint arXiv:2309.04475},
year={2023}
}
If you have any questions, feel free to reach us at:
Rui Jiao: jiaor21@mails.tsinghua.edu.cn