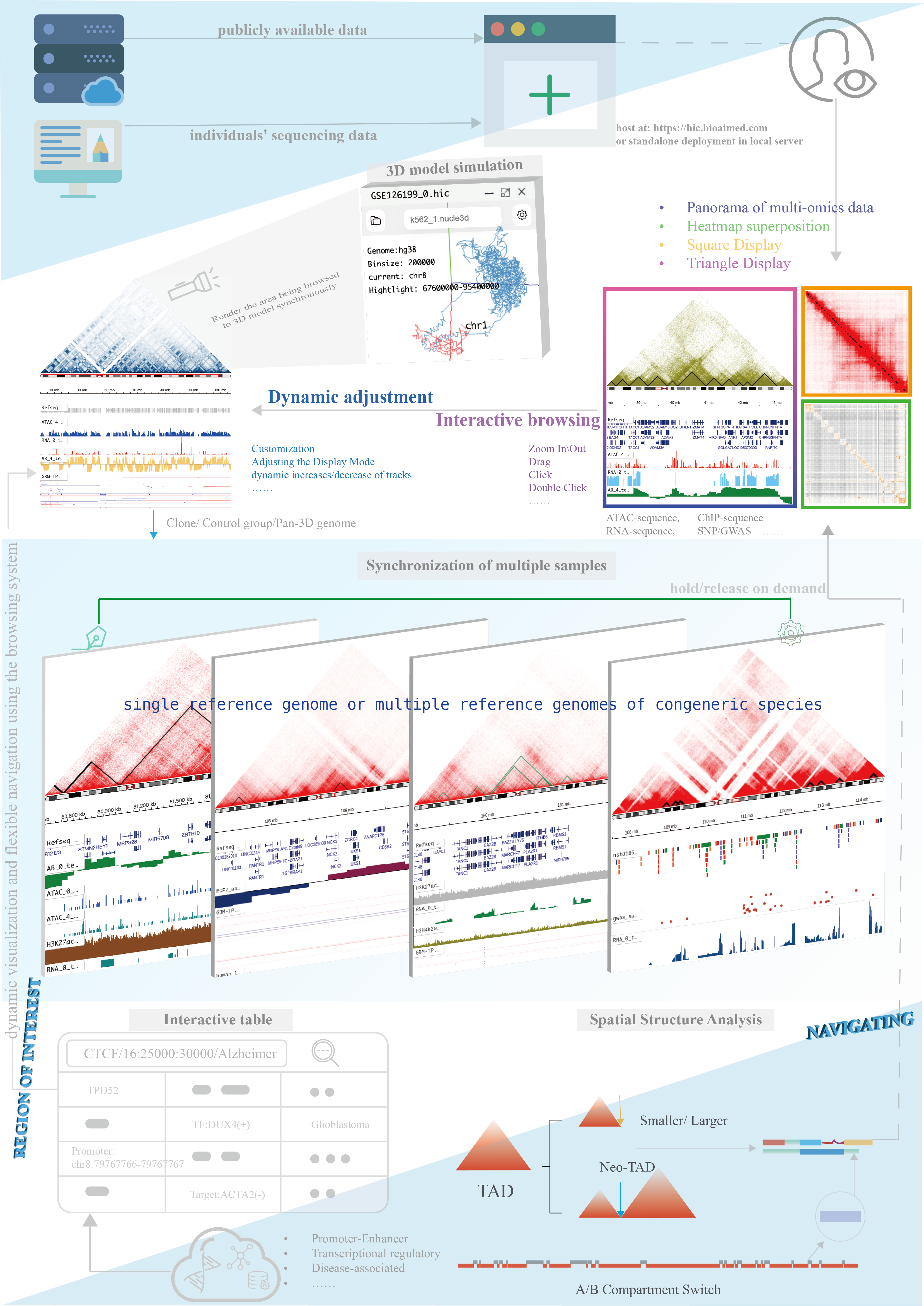
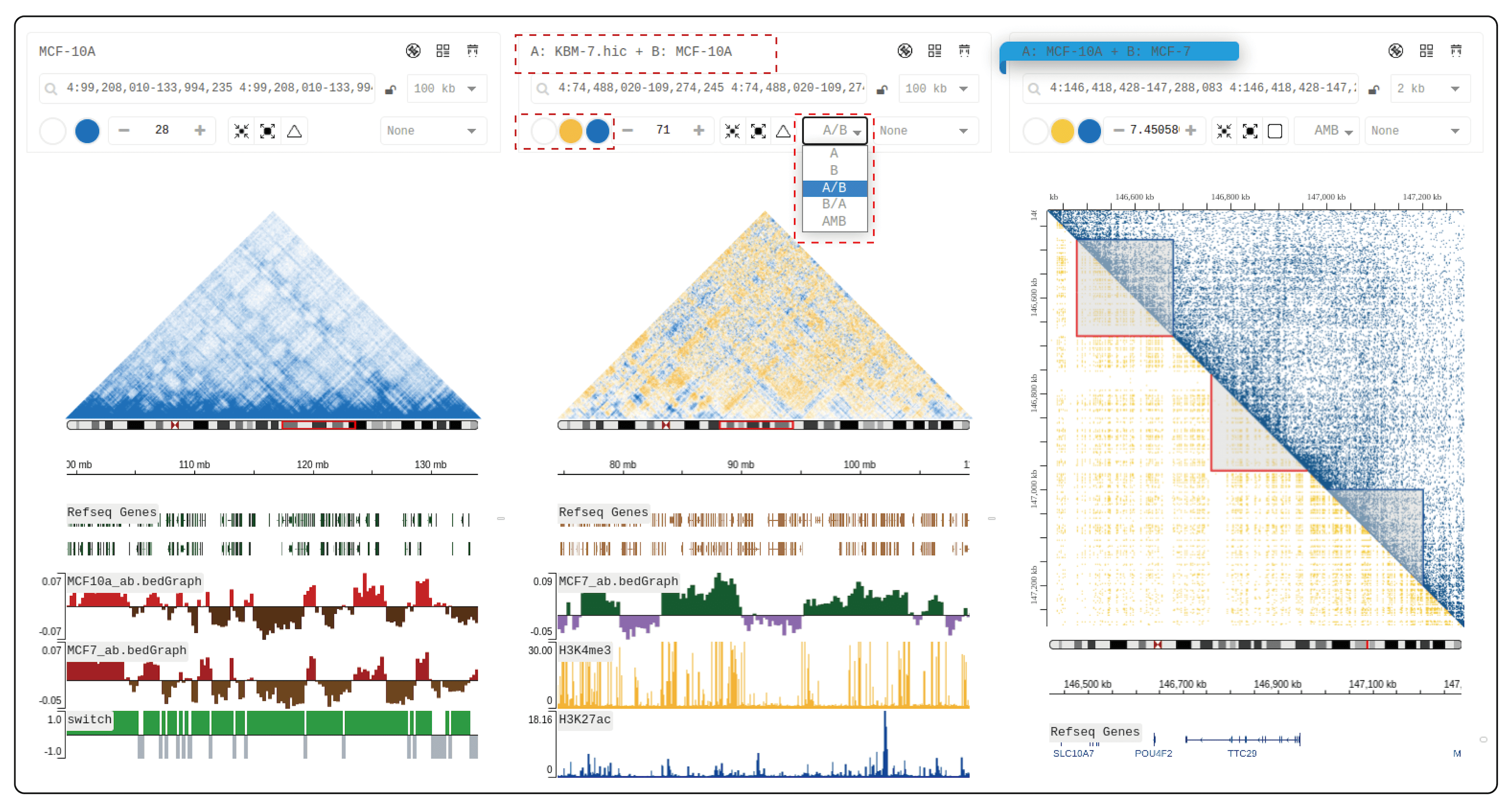
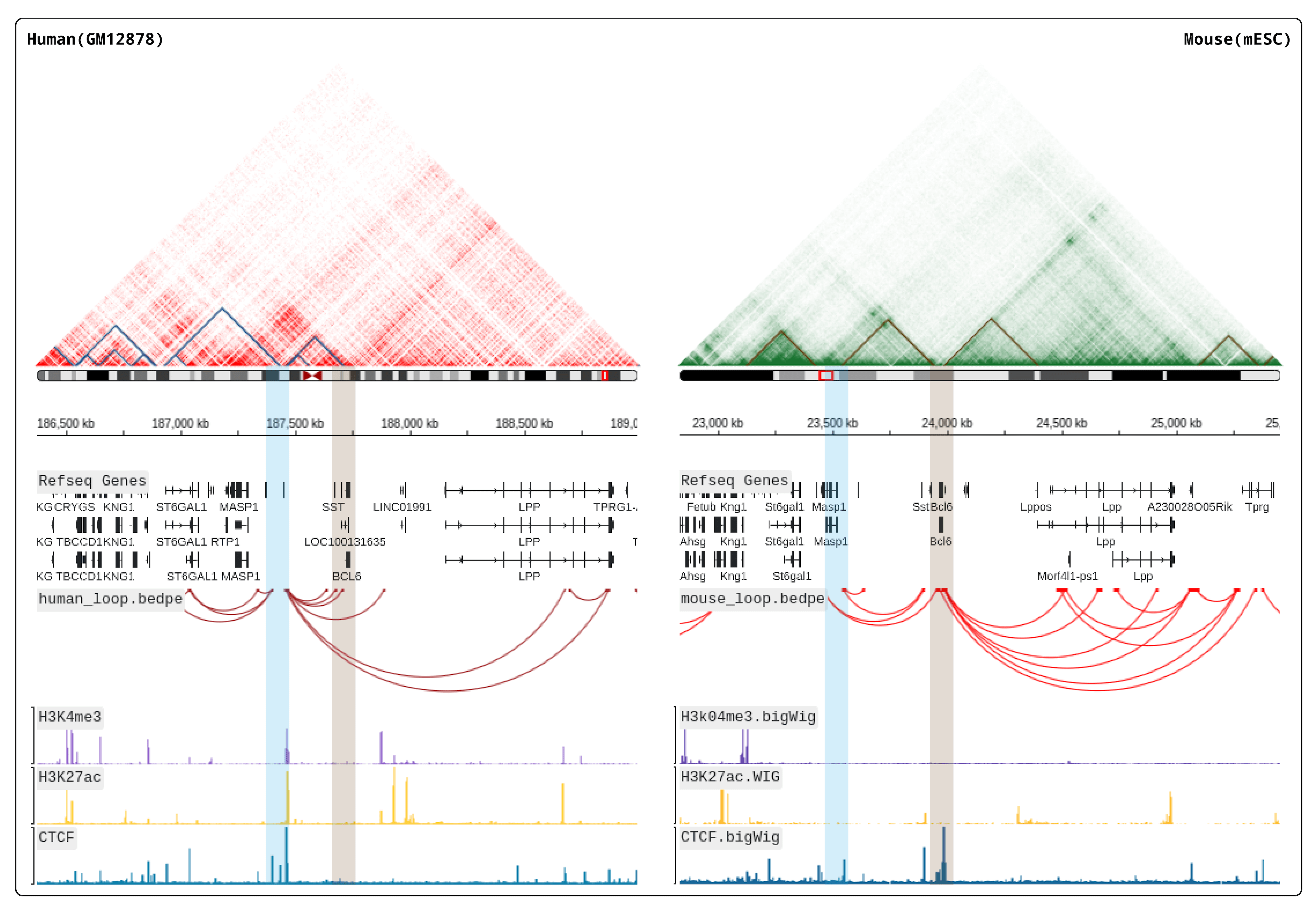
We recommend deploying HiBrowser locally so that you can access your data privatly or data in your LAN.
Although the instance we host at https://hic.bioaimed.com is an end-user application, you need to realize that your data will be flowing on the Interet [except for visualizing your local file]
Besides, multi-omics data, especially Hi-C data, is often very large. Therefor, in our host instace, the case may spend few minutes to fetching data.
Download the source code from our github repo.
- python=3.8
- numpy
- django=4.1
- django-cors-headers=3.13.0
- requests=2.28.1
We recommend using conda to create a virtual environment.
Install conda firstly.
- Enter the
backend conda create --name <your_name> --file requirements.txtconda activate <your_name>- Modify
/HicBrowser/setting.py, Change to your IP address or domain name
ALLOWED_HOSTS = ["192.168.31.196"]python manage.py runserver 0.0.0.0:<your_port>
[For Developer]
Recommended using Visual Studio Code + Live Server
[For User]
Install nginx.
- Enter
/frontend - Copy all files to the root directory of your nginx
## How to find the root directory
# Linux
cat /usr/local/nginx/conf/nginx.conf
# Windows
## In your downloaded compressed file- Modify
/js/setting.js, change to your backend address
export const api_url = '<your_ip>:<your_port>/api';- Start your nginx
# Linux
cd usr/local/nginx/sbin
./nginx
# Windows
start nginx.exevisit http://<you_ip>
- Enter
/docs - Open
index.htmlwith your browser[eg. Google Chrome]
see usermanual.pdf in repo.
Three cases are available for users to view.
<your ip>/?example=100010
<your ip>/?example=100020
<your ip>/?example=100030
################################################ an instance we host
https://hic.bioaimed.com/browser/?example=100010
https://hic.bioaimed.com/browser/?example=100020
https://hic.bioaimed.com/browser/?example=100030
The data used in the supplementary figures is listed below.
GM12878 Cell Line: https://4dn-open-data-public.s3.amazonaws.com/fourfront-webprod/wfoutput/1201682a-a223-482d-913d-3c3972b8eb65/4DNFIIRIHBR2.hic
mES Cell Line: https://cdn.bioaimed.com/mouse/mouse.hic
ChIP-seq Data: GEO accession code GSE36028
K562 and KBM-7 Cell Lines: https://data.4dnucleome.org/publications/cf0e49aa-173c-49d1-a7c7-22acbc83c064/
Here are some test data we provide.It can be visualized in HiBrowser(your locally deployed or we host instance) by using URL. But it will be slow due to the network congestion.
You can download it from our data hub. [Total 110.35 GB]
### Hi-C file
#### human
1. https://cdn.bioaimed.com/human/4DNFI1E6NJQJ.hic
2. https://cdn.bioaimed.com/human/4DNFICSTCJQZ.hic
3. https://cdn.bioaimed.com/GSE126199/GSE126199_0.hic
4. https://cdn.bioaimed.com/GSE126199/GSE126199_4.hic
5. https://cdn.bioaimed.com/human/human.hic
#### mouse
6. https://cdn.bioaimed.com/mouse/mouse.hic
### hg19
1. https://cdn.bioaimed.com/human/Homo_sapiens_assembly19.fasta
2. https://cdn.bioaimed.com/human/Homo_sapiens_assembly19.fasta.fai
3. https://cdn.bioaimed.com/human/human_cytoBandIdeo.txt
4. https://cdn.bioaimed.com/human/human_refGene.txt.gz
### mm10
1. https://cdn.bioaimed.com/mouse/Mus_musculus.GRCm39.dna.primary_assembly.fa
2. https://cdn.bioaimed.com/mouse/Mus_musculus.GRCm39.dna.primary_assembly.fa.fai
3. https://cdn.bioaimed.com/mouse/mouse_refGene.txt.gz
4. https://cdn.bioaimed.com/mouse/mouse_cytoBand.txt
### human 3D structure
1. https://cdn.bioaimed.com/human/human_domain.txt
2. https://cdn.bioaimed.com/human/human_loop.txt
3. https://cdn.bioaimed.com/human/MCF10a_domin.txt
4. https://cdn.bioaimed.com/human/MCF10a_ab.bedGraph
5. https://cdn.bioaimed.com/human/MCF7_domin.txt
6. https://cdn.bioaimed.com/human/MCF7_ab.bedGraph
### mouse 3D structure
1. https://cdn.bioaimed.com/mouse/mouse_domain.txt
2. https://cdn.bioaimed.com/mouse/mouse_loop.txt
### track file
1. https://cdn.bioaimed.com/track/annotation/Homo_sapiens.GRCh38.94.chr.gff3.gz
2. https://cdn.bioaimed.com/track/annotation/Homo_sapiens.GRCh38.94.chr.gff3.gz.tbi
--
1. https://cdn.bioaimed.com/track/auto/case1.bed.gz
2. https://cdn.bioaimed.com/track/auto/case1.bed.gz.tbi
--
1. https://cdn.bioaimed.com/track/gwas/gwas_sample.gwas
--
1. https://cdn.bioaimed.com/track/seg/GBM-TP.seg.gz
--
1. https://cdn.bioaimed.com/track/variant/nstd186.GRCh38.variant_call.vcf.gz
2. https://cdn.bioaimed.com/track/variant/nstd186.GRCh38.variant_call.vcf.gz.tbi
### ATAC-seq
1. https://cdn.bioaimed.com/GSE126199/ATAC_0_telo.bw
2. https://cdn.bioaimed.com/GSE126199/ATAC_4_telo.bw
### RNA
1. https://cdn.bioaimed.com/GSE126199/RNA_0_telo.bw
2. https://cdn.bioaimed.com/GSE126199/RNA_4_telo.bw
### other
1. https://cdn.bioaimed.com/track/bw/GSM3609942_JWH107-CutRun-35-minus-IAA-CTCF.bam.bw
2. https://cdn.bioaimed.com/track/bw/GSM3609943_JWH108-CutRun-35-plus48-IAA-CTCF.bam.bw
3. https://cdn.bioaimed.com/track/bw/GSM3609944_JWH110-CutRun-42-minus-IAA-CTCF.bam.bw
4. https://cdn.bioaimed.com/track/bw/GSM3609945_JWH111-CutRun-42-plus48-IAA-CTCF.bam.bw
##### CTCF no IAA
1. https://cdn.bioaimed.com/track/bw/GSM4636526_ATAC-SEM-CTCF-AID_no_IAA-Rep42.free.bw
2. https://cdn.bioaimed.com/track/bw/GSM4636525_ATAC-SEM-CTCF-AID_no_IAA-Rep35.free.bw
3. https://cdn.bioaimed.com/track/bw/GSM4636524_ATAC-SEM-CTCF-AID_no_IAA-Rep27.free.bigwig
##### CTCF with IAA
1. https://cdn.bioaimed.com/track/bw/GSM4636529_ATAC-SEM-CTCF-AID_with_IAA-Rep42.free.bw
2. https://cdn.bioaimed.com/track/bw/GSM4636528_ATAC-SEM-CTCF-AID_with_IAA-Rep35.free.bw
3. https://cdn.bioaimed.com/track/bw/GSM4636527_ATAC-SEM-CTCF-AID_with_IAA-Rep27.free.bigwig