SPEAR is an annotation tool for SARS-CoV-2 genomes, it provides comprehensive annotation of all protein products, in particular, Spike (S) mutations are annotated with a range of scores that provide indications of their likely effects on ACE2 binding, and likely contribution to immune escape. The aim of SPEAR is to provide a lightweight genomic surveillance tool that can be run within a diagnostic lab, sequencing facility, or analysis pipeline providing quantitative scores at point of sequencing. Functional annotation and effect scoring are derived from protein structure, theoretical simulation, and omics' experiments.
SPEAR is capable of running on single or multiple input files and accepts a range of standard inputs, FASTA consensus sequences .fa, sequences aligned to reference genome (NC_045512.2|MN908947.3) .aln and .vcf files. SPEAR will annotate and score 1,000 consensus input sequences in ~12.5 mins using a single CPU core and in ~3.5 mins with 8 cores.
The SPEAR scoring system identifies both the potential immune escape and reduced ACE2 binding consequences of variants in the Omicron RBD, as well as highlighting the potential increased stability of the open conformation of the Spike protein within Omicron, a more in-depth discussion of these matters can be found in our preprint Teruel et al[1].
SPEAR is now published in Bioinformatics, if you use SPEAR in your work please cite our paper:
Crown M, Teruel N, Najmanovich R, Basthon M. SPEAR: Systematic ProtEin AnnotatoR Bioinformatics, btac391, https://doi.org/10.1093/bioinformatics/btac391
Clone this repo:
git clone https://github.com/m-crown/SPEAR.git
Change to downloaded directory:
cd SPEAR
Run the install script, (this requires a working conda install, we recommend miniconda):
install_spear
Activate the conda environment:
conda activate spear
Run spear:
spear
Once installed SPEAR can be updated with the spear update command, where:
spear update spear- updates the core SPEAR code and SPEAR data from this repo.spear update data- updates SPEAR data and external 3rd party datasets.spear update all- updates everything and reinstalls the conda env as well as SPEAR.
SPEAR has been tested on Intel Mac OS 10.5, 11.x, 12.x, RHEL clones 7/8 and Ubuntu 21.10, and should work on other Linux distributions.
SPEAR is driven by input modality so your first argument to it should reflect the type of input files you are providing, spear consensus for analysis of .fa genome consensus files, spear alignment for analysis of pre-aligned consensuses in multiple FASTA format .aln. And spear vcf for the analysis of .vcf files. The reference genome used should be either NC_045512.2 or MN908947.3.
usage: spear [-h] {consensus,alignment,vcf,update,demo} ...
positional arguments:
{consensus,alignment,vcf,update,demo}
consensus Run SPEAR on consensus FASTA sequence (align first).
alignment Run SPEAR on alignment in FASTA format (skip alignment).
vcf Run SPEAR on existing VCF file(s) - skip alignment and SNP/indel identification and ambiguous SNP filtering.
update Update [spear,data,all]
demo Run SPEAR demo on lineage VCFs
options:
-h, --help show this help message and exit
Please select a subcommand (choose from 'consensus', 'alignment', 'vcf', 'update', 'demo')
Further options are then available depending on the type of input file, e.g. spear consensus --help:
usage: spear alignment [-h] [--debug] [--dag] [--no-report] [--tmp] [--extension]
[--mask-problem-sites AB AM HA [AB AM HA ...]] [--threads] [--aligner] [--cutoff] [--global_n]
[--s_n] [--s_contig] [--rbd_n] [--window] [--baseline_scores] [--baseline]
[--no-product-plot]
input output
positional arguments:
input Input directory of alignments, consensus fasta sequences or VCF files.
output Destination dir for SPEAR annotated VCFs
options:
-h, --help show this help message and exit
--debug Verbose snakemake execution
--dag Display DAG and exit
--no-report Do not produce HTML report
--tmp Preserve intermediate output files for debugging.
--extension Suffix and extension for input files
--mask-problem-sites AB AM HA [AB AM HA ...]
Filter problematic sites with these codes: [AB AM HA HH HO IC NA NS NL SS AD BR all]
--threads Max number of threads for snakemake job execution.
--aligner Alignment method to use for alignment to SARS-CoV-2 reference, 'minimap2' or 'muscle', default minimap2
--cutoff Percentage N cutoff for input sequences. Default 50
--global_n Minimum percentage of N in sample to flag as poor coverage. Default half of cutoff.
--s_n Minimum percentage of N in S gene to flag as poor coverage. Default 5.
--s_contig Minimum length of contig to flag sample as potential S gene dropout. Default 150nt
--rbd_n Number of N's in sample spike RBD to flag as poor. Default 12nt
--window Maximum number of flanking N's around deletion, default 2
--baseline_scores Custom baseline scores file for use in summary report
--baseline Baseline sample to use, either from pre-loaded baseline scores or user-supplied custom
baseline file. Default BA.2. Built-in options: BA.1 BA.1.1 BA.2 Omicron Delta Alpha
--no-product-plot Do not produce individual sample product plots (for fastest operation)
--pangolin PANGOLIN Pangolin operation mode: accurate (UShER), fast (pangolearn), none (don't run pangolin)
To check installation was successful and view an example SPEAR run and report, run the built-in demo:
spear demo
This will save output including the HTML report to the demo_out/ directory within your current working directory.
To use spear on a single .fa consensus file:
spear consensus sample1.fa output
This will launch spear analyse sample1.fa and write the output to a directory tree contained within output/.
To run on multiple input files replace the input file name with a directory:
spear consensus consensus_files output
You can also use . as input directory to use files in the current working directory.
By default consensus files are assumed to have the extension .fa, alignments .aln and vcf files .vcf, if you have a different extension then specify the suffix with --extension. This also allows you to remove a suffix from the sample ID used in output, so if all your input alignments conform to <sample_id>.muscle.aln specifying: --extension .muscle.aln will ensure only the sample id/name makes it into the output. Note that running on multiple input files may require you to increase the maximum number of open file handles on your system if your number of input samples starts to approach this limit, check this with ulimit -n.
Consensus inputs can be aligned to reference using either MUSCLE v3.8 or minimap2, specified using --aligner. From version 0.8 onwards the default alignment method is minimap2, due to the significant speed improvements - 15.7X speedup on single thread, 9.9X speedup on quad thread and 8.8X speedup on eight threads. Users should be aware that small differences, particularly in resolution of indels can occur between MUSCLE and minimap2 alignments where the alignment solution is ambiguous - these cases are rare, and the vast majority of alignments should agree.
If using spear alignment please make sure your samples are aligned to NC_045512.2 or MN908947.3, and that the multiple FASTA format is used, (expected file extension .aln) with the reference sequence being the fist one within the alignment.
Make sure you VCF file uses NC_045512.2 or MN908947.3 as its reference and that all variants are co-ordinate sorted.
By default SPEAR will filter out the variants occurring in "genomic scraggly ends" the very most 5' 1-55 nucleotides and final 3' end of the genome 29,804-29,903. Input consensus FASTA will also be filtered to exclude samples that are more than 50% N before they are aligned to the reference. Percentage N filtering can be tuned with --cutoff. Further filtering options are discussed in Advanced filtering options below.
Pangolin lineage assignment will be run by default on all samples, (including VCF), this enables grouping of sample by lineage in the output report. There are three pangolin operation modes in spear: accurate (UShER), fast (pangolearn), none (don't run pangolin). Please note that with VCF inputs consensus fasta sequences will be reconstructed from VCF and all other bases are assumed to be reference. We recommend conducting appropriate QC outside of SPEAR on these samples. Where possible please use sequence inputs so that SPEAR can assess dropouts, %N, (See QC section below) and pass ambiguous base calls to pangolin.
All spear output is nested into the output directory specified at run time.
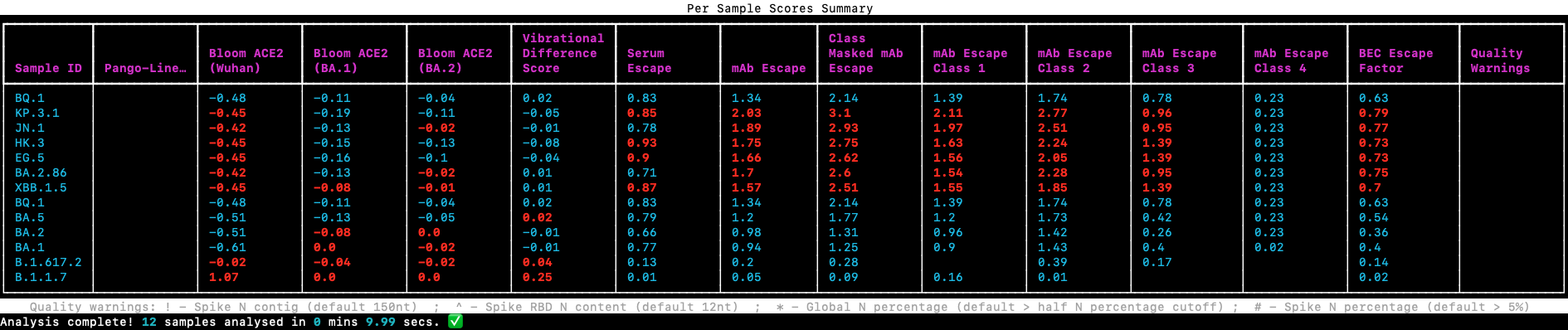
Scores for each sample along with highlights showing where these exceed the chosen baseline (defaults to BA.2) are produced in the SPEAR terminal output. This mirrors the SPEAR Per Sample Scores Summary table found in the HTML report. And is sorted by the column Class Masked mAb escape.
All scores (as discussed below) can be compared to a baseline lineage or user-supplied sample, the default is to compare to BA.2, this means that any scores above the values in the baseline will be highlighted, potentially flagging samples with enhanced immune escape or ACE2 binding, these scores are discussed below. To select an alternative baseline from the built-in options use:
--baseline BA.1 or BA.1.1, BA.2, Omicron, Delta, Alpha
A user supplied sample can be used as baseline by specifying both a spear_score_summary.tsv file and the sample_id:
--baseline_scores spear_score_summary.tsv --baseline sample_id
Baseline lineages VCFs, their composition and creation are discussed in SPEAR-Reports.
SPEAR will quality check and flag issues within any consensus or alignment inputs for: global N percentage (>25%), Spike N percentage (>5%), Spike dropout (contig of N >150nt), and Receptor Binding Domain (RBD) quality (>12nt N content). These QC checks do not work for VCF input. Dropout detection is critical as missing mutations in Spike can’t be scored. These QC warnings are displayed in the final column of the terminal output and Per Sample Score Summary table: ! - Spike N contig, ^ - Spike RBD N content, * - Global N percentage, # - Spike N percentage. These values are user configurable. The end user should always check the quality of input data and underlying variant calls, as whilst steps are taken to alert the user to issues with input sequences these don't replace robust sequencing QC.
all_samples.spear.vcf- a multi sample VCF file with all annotations encoded in VCF format, header describes SPEAR fields.spear_annotation_summary.tsv- a tab delimited file for all samples with SPEAR annotation and scores per variant, one row per variant.spear_score_summary.tsv- a tab delimited file with total scores for each sample, one sample per row.spear_variant_summary.csv- a comma delimited file, one row per sample listing all variants and their consequence type.qc.csv- a comma delimited file with QC data, columns:sample_id,global_n(global N%),s_n(Spike N%),s_n_contig(longest Spike contig of N),rbd_n(RBD N nt count).report/- this directory will contain an HTMLreport.htmlsupporting files are also required within this directory tree.report/plots/- this directory contains all standalone plots also found in the above report.lineage_report.csv- output from Pangolin (if Pangolin is not run this file will be empty).
final_vcfs/- within this directory there will be a VCF file per sample, SPEAR fields format is as all sample file.per_sample_annotation/- within this directory there will be a tab delimited file per sample with all SPEAR annotation and scores.reports/plots/product_plots/- this directory contains a protein product plot per sample with individual mutations shown along with scores where appropriate.
This is an interactive report, with sortable tables of variants, residues, scores, and interactive heatmaps of scores for the Spike protein. An example report can be found here. ORF plots for each individual samples showing mutations placed on to ORFs/products are also found at the bottom of this report.
These columns are within both the per sample files and spear_annotation_summary.tsv:
| Column ID | Description |
|---|---|
sample_id |
Input sample ID, taken from input file header in .fa, .aln and sample col in .vcf. |
POS |
Position of variant as per VCF spec (1-based). |
REF |
Reference genome base(s) as per VCF spec. |
ALT |
Alternative base(s) as per VCF spec. |
gene_name |
ORF1ab, S, ORF3a, E, M, ORF6, ORF7a, ORF7b, ORF8, N, variants between ORFs are flagged as intergenic. |
HGVS.nt |
HGVS annotation for c. coding regions, or n. non-coding with nucleotide co-ordinates, note our HGVS isa not 3' aligned as per HGVS standard, as this causes issues with incorrect consequence calling, as such all positions honour original VCF co-ordinates. |
consequence_type |
The type of variant consequence, e.g. missense_variant, synonymous_variant, multiple consequences are possible here and will be & separated. |
description |
Free text description of product, ORF1ab is further broken down here into: leader protein (nsp1), nsp2, nsp3, nsp4, 3C-like proteinase (nsp5), nsp6, nsp7, nsp8, nsp9, nsp10, RNA-dependent RNA polymerase (RdRp, nsp12), helicase (nsp13), 3'-to-5' exonuclease (nsp14), endoRNAse (nsp15), and 2'-O-ribose methyltransferase (nsp16). |
RefSeq_acc |
RefSeq accession for protein product, will be final cleaved polypotein product specific. |
residues |
Amino acid residue changes in the format of N501Y (REF-AA residue ALT-AA), where the genomic events produces multiple changes these will be expressed like so: G142D,del143,del144,del145 for G142D followed by deletion of 3 residues, or R203K,G204R exposing individual AA changes within a MNP. |
region |
Currently only annotated for Spike: S1 or S2 and ORF3a transmembrane or cytosolic. |
domain |
Annotated for protein where multiple domains are present, spike definitions, e.g. NTD, RBD, RBM, definitions can be found here. |
feature |
Features possessed by residues distinct from domain annotation, such as active sites or ligand binding roles where each annotated residue contributes to the feature. |
contact_type |
Takes the format for molecule:bond_type_residue, e.g. ACE2:h-bond_E35+contact_K31_H34, implies a ACE2 h-bond made by annotated residue to E35 within ACE2 as well as 4Å cut-off contact made to K31 and H34, + delimits additional contact types. trimer:h-bond_707_709+contact implies a contact in the trimer interface of Spike h-binding to residues 707 and 709 with an additional generic none residue specific contact. The bond type salt-bridge is also possible here. Currently only annotated for Spike. |
NAb |
A list of bound neutralising antibodies, this list is + delimited, with , reserved to delimit multiple amino acid variant events as described in residues. Currently only annotated for Spike. |
barnes_class |
If the residue is part of a Barnes epitope class as defined in Barnes et al.[2], annotated values are 1, 2, 3, 4 with a + delimiter, some classes are appended with a * where they were not part of formal epitope studies but that residue was found to be sensitive to biding of antibodies of that class via mutagenesis studies. |
Table 1. Per sample annotation.
The next 12 columns of output are scores rather than annotations and are described below.
SPEAR uses a number of different scores to evaluate the likely impact of viral genomes, these can be found at the residue level in the .spear.annotation.tsv files within per_sample_annotation/ directory and also in the spear_annotation_summary.tsv file for all samples in the run. Some scores operate at a per residue level, such that any variant will get the same score, while others are mutation specific (accounting for individual amino acid change), and some operate at a whole sample level.
| Column ID | Level | Description |
|---|---|---|
bloom_ace2_wuhan |
mutation | ACE2 binding value Δ-log10(KD,app) relative to the "wild-type" (WT), data from Starr et al.[3] Higher positive values mean binding is stronger than WT, negative values mean binding is weaker than WT. |
bloom_ace2_BA1 |
mutation | ACE2 binding value Δ-log10(KD,app) relative to BA.1. Higher positive values mean binding is stronger than WT, negative values mean binding is weaker than BA.1. |
bloom_ace2_BA2 |
mutation | ACE2 binding value Δ-log10(KD,app) relative to BA.2. Higher positive values mean binding is stronger than WT, negative values mean binding is weaker than BA.2. |
VDS |
mutation | Vibrational Difference Score (VDS), positive VDS values suggests mutation stabilises the open state of Spike and/or makes the closed state more flexible, favouring the open conformation relative to the WT. Negative values suggest mutation favours the closed state more than WT. Data from Teruel et al.[4]. |
serum_escape |
mutation | Mean residue specific serum escape score from 7 individuals in Greaney et al.[5], larger values indicate more escape, (range 0-1). |
mAb_escape |
mutation | Mean residue specific mAb escape score from 26 mAbs, larger values indicate more escape, (range 0-1). Data taken from Dong et al.[6], SARS-CoV-2-RBD_MAP_COV2-2955[7], Greaney et al.[8], Starr et al.[9], Starr et al.[10], Starr et al.[11] Tortorici et al.[12]. |
cm_mAb_escape |
mutation | As above, but calculated in a Barnes class mask specific way such that the mean is taken only from Barnes class mAbs that correspond to class of residue with mutation. |
mAb_escape_class_1 |
mutation | As above, mean residue specific mAb escape score from class 1 mAbs only, only applied to residues in Barnes class 1 epitope. |
mAb_escape_class_2 |
mutation | As above, mean residue specific mAb escape score from class 2 mAbs only, only applied to residues in Barnes class 2 epitope. |
mAb_escape_class_3 |
mutation | As above, mean residue specific mAb escape score from class 3 mAbs only, only applied to residues in Barnes class 3 epitope. |
mAb_escape_class_4 |
mutation | As above, mean residue specific mAb escape score from class 4 mAbs only, only applied to residues in Barnes class 4 epitope. |
BEC_RES |
residue | Bloom Escape Calculator Residue Escape Score, this residue specific number is generated from the full compliment of mutated residues in the sample using bindingcalculator.py from jbloomlab/SARS2_RBD_Ab_escape_maps as described in Greaney et al.[13]. Lower values here indicate more antibody escape. |
BEC_EF |
residue | Bloom Escape Calculator Escape Factor, a fraction (0 to 1) of antibodies escaped by mutations at this residue. 0 = no antibodies escaped, 1 = all antibodies escaped. This value is calculated for individual mutations without contribution of other mutated residues. |
BEC_sample_EF |
sample | Bloom Escape Calculator Escape Factor as BEC_EF but calculated using the full compliment of mutated residues in the sample, this score will be the same for every mutated residue in a sample. |
Table 2. Per sample scores.
These scores are also summarised in spear_score_summary.tsv with a row for each sample. Some columns summarise values for multiple entities here and are internally , delimited. Documentation for this file is found in Table 4.
Some of the scores employed here have been used to demonstrate the immune escape and ACE2 binding properties of Omicron and are discussed further in Teruel et al. [1].
There are known problematic sites in SARS-CoV-2 sequencing, these sites are not filtered out by default, and their usages needs to be handled with caution. This list is maintained for a tree building usage case, and sites such as S:G142D which are informative from an annotation perspective may be masked if this list is used blindly, owing to the fact that this region is difficult to call on ARTIC v3 and other primers in Delta genomes. Thus considerations for building phylogenetic trees are not always ideal from an annotation perspective. To get round this we expose the ability to filter sites flagged as "mask" with user defined granularity by invoking --mask-problem-sites along with one or more of the following two letter codes which indicate which of the tags described originally here and reproduced below will be filtered:
| Code | Tag | Description |
|---|---|---|
| AB | ambiguous | Sites which show an excess of ambiguous basecalls relative to the number of alternative alleles, often emerging from a single country or sequencing laboratory. |
| AM | amended | Previous sequencing errors which now appear to have been fixed in the latest versions of the GISAID sequences, at least in sequences from some of the sequencing laboratories. |
| HA | highly_ambiguous | Sites with a very high proportion of ambiguous characters, relative to the number of alternative alleles. |
| HH | highly_homoplasic | Positions which are extremely homoplasic - it is sometimes not necessarily clear if these are hypermutable sites or sequencing artefacts. |
| HO | homoplasic | Homoplasic sites, with many mutation events needed to explain a relatively small alternative allele count. |
| IC | interspecific_contamination | Cases (so far only one instance) in which the known sequencing issue is due to contamination from genetic material that does not have SARS-CoV-2 origin. |
| NA | nanopore_adapter | Cases in which the known sequencing issue is due to the adapter sequences in nanopore reads. |
| NS | narrow_src | Variants which are found in sequences from only a few sequencing labs (usually two or three), possibly as a consequence of the same artefact reproduced independently. |
| NL | neighbour_linked | Proximal variants displaying near perfect linkage. |
| SS | single_src | Only observed in samples from a single laboratory. |
| AD | amplicon_drop_or_primer_artefact | Amplicon dropout and/or failed primer trimming. |
| BR | back_to_ref | The alternate allele is sometimes not called for this position due to issues with amplicon dropout and/or primer trimming. |
| all | all of the above | Everything marked as mask. |
Table 3. Filtering codes
All sites flagged within W-L/ProblematicSites_SARS-CoV2 will be annotated within the SPEAR output .vcf
Primary SPEAR development is undertaken by Matthew Crown project is led by Matthew Bashton and is developed in collaboration with the Najmanovich Research Group specifically Natália Teruel and Rafael Najmanovich. This work is funded by COG-UK.
Spear makes use of the following:
- conda
- bioconda Team et al.[14]
- snakemake Mölder et al.[15]
- muscle Edgar [16]
- bcftools Danecek et al.[17]
- SnpEff and SnpSift Cingolani et al.[18],Cingolani et al.[19]
- UCSC faToVCF
- vcfanno Pedersen et al.[20]
- Binding Calculator Greaney et al.[13]
- Minimap2 Heng Li [21]
- gofasta
- pangolin
- Plotly
- Bootstrap
- Rich
- Teruel, N., Crown, M., Bashton, M. & Najmanovich, R. Computational analysis of the effect of SARS-CoV-2 variant Omicron Spike protein mutations on dynamics, ACE2 binding and propensity for immune escape. Biorxiv 2021.12.14.472622 (2021) doi:10.1101/2021.12.14.472622.
- Barnes, C. O. et al. Structures of Human Antibodies Bound to SARS-CoV-2 Spike Reveal Common Epitopes and Recurrent Features of Antibodies. Cell 182, 828-842.e16 (2020).
- Starr, T. N. et al. Deep Mutational Scanning of SARS-CoV-2 Receptor Binding Domain Reveals Constraints on Folding and ACE2 Binding. Cell 182, 1295-1310.e20 (2020).
- Teruel, N., Mailhot, O. & Najmanovich, R. J. Modelling conformational state dynamics and its role on infection for SARS-CoV-2 Spike protein variants. Plos Comput Biol 17, e1009286 (2021).
- Greaney, A. J. et al. Comprehensive mapping of mutations in the SARS-CoV-2 receptor-binding domain that affect recognition by polyclonal human plasma antibodies. Cell Host Microbe 29, 463-476.e6 (2021).
- Dong, J. et al. Genetic and structural basis for SARS-CoV-2 variant neutralization by a two-antibody cocktail. Nat Microbiol 6, 1233–1244 (2021).
- https://github.com/jbloomlab/SARS-CoV-2-RBD_MAP_COV2-2955
- Greaney, A. J. et al. Mapping mutations to the SARS-CoV-2 RBD that escape binding by different classes of antibodies. Nat Commun 12, 4196 (2021).
- Starr, T. N., Greaney, A. J., Dingens, A. S. & Bloom, J. D. Complete map of SARS-CoV-2 RBD mutations that escape the monoclonal antibody LY-CoV555 and its cocktail with LY-CoV016. Cell Reports Medicine 2, 100255 (2021).
- Starr, T. N. et al. Prospective mapping of viral mutations that escape antibodies used to treat COVID-19. Science 371, 850–854 (2021).
- Starr, T. N. et al. SARS-CoV-2 RBD antibodies that maximize breadth and resistance to escape. Nature 597, 97–102 (2021).
- Tortorici, M. A. et al. Broad sarbecovirus neutralization by a human monoclonal antibody. Nature 597, 103–108 (2021).
- Greaney, A. J., Starr, T. N. & Bloom, J. D. An antibody-escape calculator for mutations to the SARS-CoV-2 receptor-binding domain. Biorxiv 2021.12.04.471236 (2021) doi:10.1101/2021.12.04.471236.
- Team, T. B. et al. Bioconda: sustainable and comprehensive software distribution for the life sciences. Nat Methods 15, 475–476 (2018).
- Mölder, F. et al. Sustainable data analysis with Snakemake. F1000research 10, 33 (2021).
- Edgar, R. C. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. Bmc Bioinformatics 5, 113 (2004).
- Danecek, P. et al. Twelve years of SAMtools and BCFtools. Gigascience 10, giab008 (2021).
- Cingolani, P. et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 6, 80–92 (2012).
- Cingolani, P. et al. Using Drosophila melanogaster as a Model for Genotoxic Chemical Mutational Studies with a New Program, SnpSift. Frontiers Genetics 3, 35 (2012).
- Pedersen, B. S., Layer, R. M. & Quinlan, A. R. Vcfanno: fast, flexible annotation of genetic variants. Genome Biol 17, 118 (2016).
- Heng, L. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34, 3094-3100 (2018).