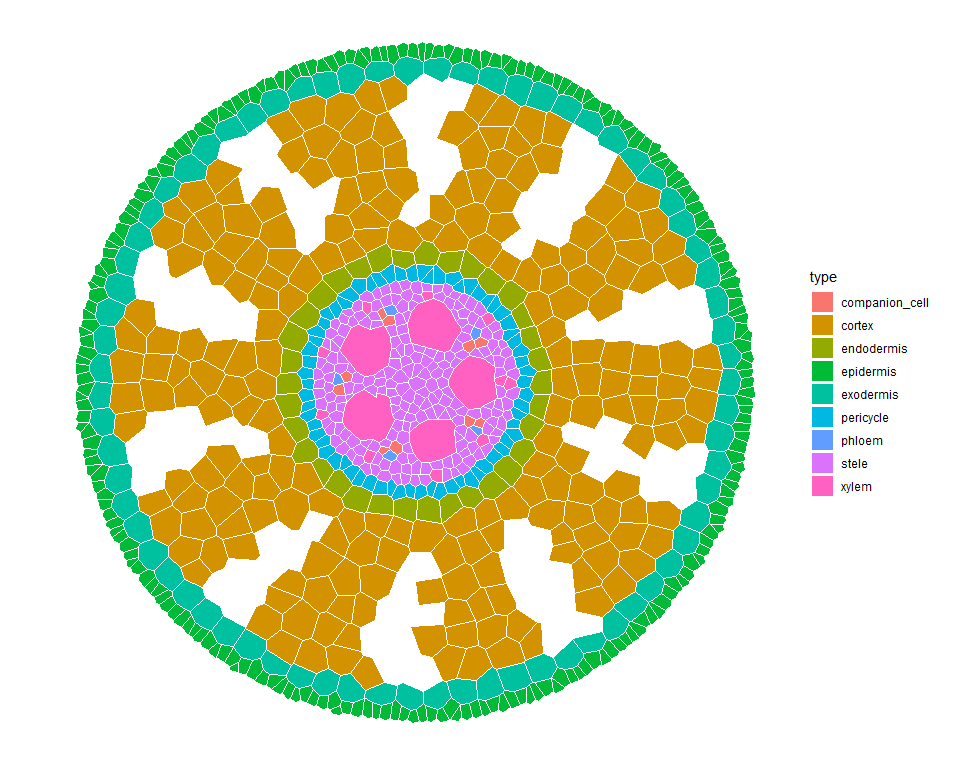
What is GRANAR?
GRANAR stand for Generator of Root ANAtomy in R. The GRANAR model is able to generate complete cell networks of a root cross-sections using a small set of root anatomical features. The root anatomical features can typically be gathered by using software image analysis such as ImageJ. Once the root anatomy is created, the cell network can be saved as an eXtended Markup Language (XML) file. The structure of the XML files created by GRANAR are identical to the ones created of CellSeT (a software which digitize root anatomical network).
GRANAR is an R package. The 'granar' package can be found on GitHub. Alternatively, by simply executing the following line in the R environment:
install.packages("devtools")
library(devtools)
install_github("granar/grarar")
library(granar)
# Load the libraries
library(xml2)
library(tidyverse)
library(plyr)
library(deldir)
library(alphahull)
library(viridis)
library(cowplot)
library(retistruct)
library(Hmisc)
# Read the parameters
params <- read_param_xml("Zea_mays_1_Heymans_2019.xml")
# Run the model
sim <- create_anatomy(parameters = params)
# plot the output
plot_anatomy(sim, col = "type")