Pytorch implementation of Bandit Optimization algorithms for materials exploration.
Details can be found in the paper to be submitted to NeurIPS 2021 Workshop AI for Science
(https://openreview.net/forum?id=xJhjehqjQeB)
Two-stage learning.
- Self-Supervised Representation Learning using CGCNN (https://github.com/txie-93/cgcnn)
- Bandit Optimization using Thompson Sampling
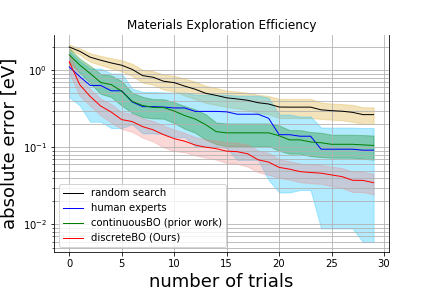
Explore the materials as close to 2.534 eV indirect bandgap as possible from 6,218 candidates with 100 prior information.
Solid lines and painted regions represent mean values and 95% confidence intervals, respectively.
Prior work based on continuous Bayesian optimization cannot explore better materials at the 30th trial than human experts.
Ours won 80% of human experts.
- Python 3.6
- PyTorch 1.4
- pymatgen
- scikit-learn
- GPyTorch
- BoTorch
- Ax-platform
Open human_experiment.ipynb, then run each cell.
Explore materials exploiting information, such as lattice parameters, chemical formula, electronegativity.
Do it 30 times within 60 mins, then generated trial_result.csv is your result.
You can compare your exploration efficiency with the model.
Run run_bandit.py
This model will explore the 6,218 candidates using 12-dim descriptors extracted through CGCNN.
The 12-dim descriptors are already converted and saved in descriptors.csv in the database folder.
Therefore, you don't need to run self-supervised representation learning.
If you want to convert cif files into descriptors, you can use pretraied model.
- go into cgcnn directory
- unzip cif.zip
- run extract.py to convert cif files into 128-dim descriptors using pretrained model (model_best.pth.tar)
- run pca.py to reduce 128-dim into 12 dim descriptors.
Masaki Adachi, High-Dimensional Discrete Bayesian Optimization with Self-Supervised Representation Learning for Data-Efficient Materials Exploration, NeurIPS Workshop AI for Science 2021