This code is based entirely on OpenMM's documentation:
Example of a ligand pose before and after energy minimisation. The pose before minimzation is shown in white, the optimised prediction is shown in pink, and the crystal ligand is shown as reference in light blue. Note how the aromatic rings are flattened and the leftmost bond is shortened by the optimisation. 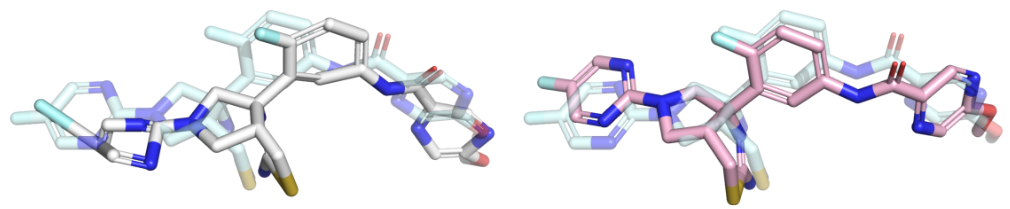
git clone https://github.com/maabuu/posebusters_em.git
cd posebusters_em
conda create -n posebusters_em openff-forcefields openff-interchange openff-toolkit "openmm>=8" "openmmforcefields>=0.11" pdbfixer "rdkit>=2022" -c conda-forge
conda activate posebusters_em
python energy_minimization.py test_cases/7MYU_ZR7/7MYU_ZR7_protein.pdb test_cases/7MYU_ZR7/7MYU_ZR7_prediction.sdf 7MYU_ZR7_prediction_minimized.sdf -t cache_dir
This example takes ~2 minutes to run on a desktop computer. Depending on the system's complexity, runs may take much longer.