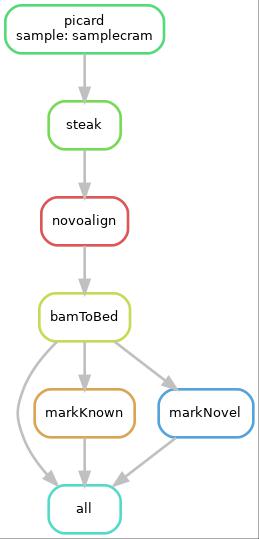
This is a SnakeMake pipeline which runs STEAK, a tool for discovery of transposable element variants. It starts with cram to bam conversion if needed, sorting, running STEAK, processing STEAK results to split prediction in novel and known.
This pipeline requires installation of snakemake pipeline management system, which is easily done by following the instructions here: https://snakemake.readthedocs.io/en/stable/getting_started/installation.html
It also needs installation of STEAK: https://github.com/applevir/STEAK
All other dependencied are automatically installed at runtime if they are not present.
If you are running this on Rosalind, STEAK module is already installed and loaded.
If you already have a reference genome (and it is indexed), update the path to it in config.yaml If you do not, run the script downloadHG19.sh in order to install it and index it.
bash downloadHG19.sh PATH_TO_DIR_TO_PLACE_REFERENCE_GENOME
update the path to the newly added and indexed human genome in config.yaml
Once the paths in config.yaml point to CRAM/BAM path, path to the reference genome and the output path - you can run the pipeline on the provided sample.bam in order to predict insertions and split them in known and novel.
snakemake --use-conda --use-envmodules --cores 1 <MY_OUTPUT_DIRECTORY>/{known/sample.knownHits.bed,novel/sample.novelHits.bed}
This will produce the two result files: <MY_OUTPUT_DIRECTORY>/known/sample.knownHits.bed: a bed file with known insertions <MY_OUTPUT_DIRECTORY>/novel/sample.novelHits.bed: a bed file with novel insertions