In this solution accelerator, we will build a Common Data Model for bservational research based on OMOP 5.31 CDM, on databricks lakehouse platform.
Observational databases are designed to primarily empower longitudinal studies and differ in both purpose and design from Electronic Medical Records (aimed at supporting clinical practice at the point of care), or claims data that are built for the insurance reimbursement processes. The Common Data Model is designed to address this problem:
The Common Data Model (CDM) can accommodate both administrative claims and EHR, allowing users to generate evidence from a wide variety of sources. It would also support collaborative research across data sources both within and outside the United States, in addition to being manageable for data owners and useful for data users.
One of the most widely used CDMs for observational research is The OMOP Common Data Model.
The OMOP Common Data Model allows for the systematic analysis of disparate observational databases. The concept behind this approach is to transform data contained within those databases into a common format (data model) as well as a common representation (terminologies, vocabularies, coding schemes), and then perform systematic analyses using a library of standard analytic routines that have been written based on the common format.
for more information visit https://www.ohdsi.org/data-standardization/the-common-data-model/
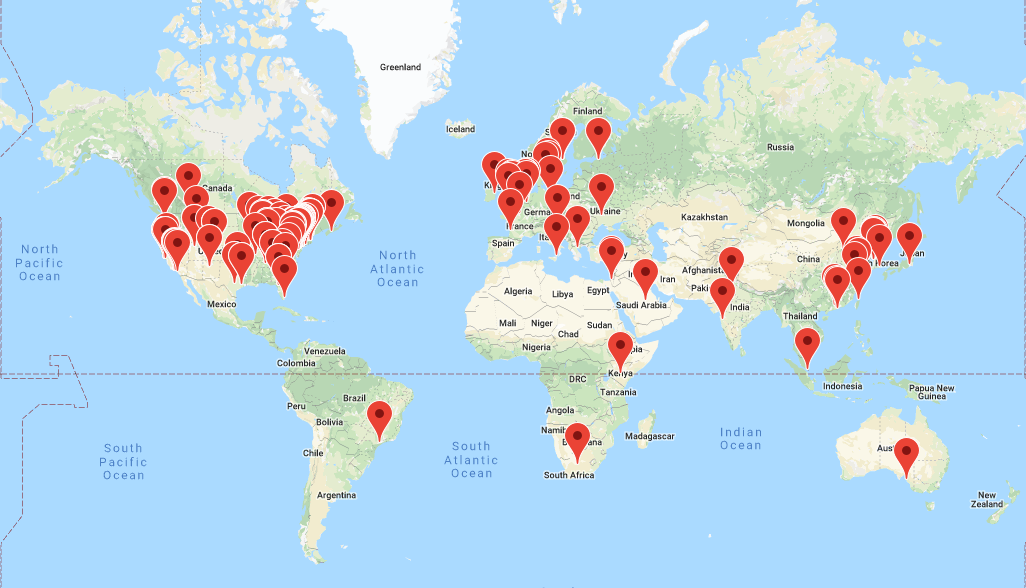
OMOP CDM has now been adopted by more than 30 countries with more than 600M unique patient records.
The complexity of such healthcare data, data variety, volume and varied data quality, pose computational and organizational challenges. For example, performing cohort studies on longitudinal data requires complex ETL of the raw data to transform the data into a data model that is optimized for such studies and then to perform statistical analysis on the resulting cohorts. Each step of this process currently requires using different technologies for ETL, storage, governance and analysis of the data. Each additional technology adds a new layer of complexity to the process, which reduces efficiency and inhibits collaboration.
The lakehouse paradigm, addresses such complexities: It is an architecture that by design brings the best of the two worlds of data warehousing and data lakes together: Data lakes meet the top requirement for healthcare and life sciences organizations to store and manage different data modalities and allow advanced analytics methods to be performed directly on the data where it sits in the cloud. However, datalakes lack the ability for regulatory-grade audits and data versioning. In contrast, data warehouses, which allow for quick access to data and support simple prescriptive analytics (such as simple aggregate statistics on the data), do not support unstructured data nor perform advanced analytics and ML. The lakehouse architecture allows organizations to perform descriptive and predictive analytics on the vast amounts of healthcare data directly on cloud storage where data sits while ensuring regulatory-grade compliance and security.
In this solution accelerator, we provide a few examples to show how the lakehouse paradigm can be leveraged to support OMOP CDM.
README: This notebookRUNME: Automating the end-to-end workflow0-config: configuration and setup1-data-ingest: To ingest synthetic records in csv format from cloud to delta bronze layer and create a synthea database2_omop531_cdm_setup: Definitions of OMOP 5.3.1 common data model for delta (DDL)3-omop_vocab_setup: create OMOP vocabulary tables4_omop531_etl_synthea: Example ETL for transforming synthea resources into OMOP 5.3.15-analysis/CHF-cohort-building: Example notebook to create a cohort of patients with Chronic Heart Failure and look at emergency room visit trends by gender and age5-analysis/drug-analysis: Examples for analysis of drug prescription trends5-analysis/sample-omop_queries: Sample SQL queries from OHDSI
To simulate health records, we used Synthea to generate ~90K synthetic patients, from across the US. You can also access sample raw csv files in /databricks-datasets/ehr/rwe/csv for 10K patients.
By running ../1-data-ingest you create the brozne tables based on the pre-simulated synthetic records.
© 2022 Databricks, Inc. All rights reserved. The source in this notebook is provided subject to the Databricks License [https://databricks.com/db-license-source]. All included or referenced third party libraries are subject to the licenses set forth below.
| Library Name | Library License | Library License URL | Library Source URL |
|---|---|---|---|
| Smolder | Apache-2.0 License | https://github.com/databrickslabs/smolder | https://github.com/databrickslabs/smolder/blob/master/LICENSE |
| Synthea | Apache License 2.0 | https://github.com/synthetichealth/synthea/blob/master/LICENSE | https://github.com/synthetichealth/synthea |
| OHDSI/CommonDataModel | Apache License 2.0 | https://github.com/OHDSI/CommonDataModel/blob/master/LICENSE | https://github.com/OHDSI/CommonDataModel |
| OHDSI/ETL-Synthea | Apache License 2.0 | https://github.com/OHDSI/ETL-Synthea/blob/master/LICENSE | https://github.com/OHDSI/ETL-Synthea |
| OHDSI/OMOP-Queries | https://github.com/OHDSI/OMOP-Queries | ||
| The Book of OHDSI | Creative Commons Zero v1.0 Universal license. | https://ohdsi.github.io/TheBookOfOhdsi/index.html#license | https://ohdsi.github.io/TheBookOfOhdsi/ |
To run this accelerator, clone this repo into a Databricks workspace. Attach the RUNME notebook to any cluster running a DBR 11.0 or later runtime, and execute the notebook via Run-All. A multi-step-job describing the accelerator pipeline will be created, and the link will be provided. Execute the multi-step-job to see how the pipeline runs.
The job configuration is written in the RUNME notebook in json format. The cost associated with running the accelerator is the user's responsibility.