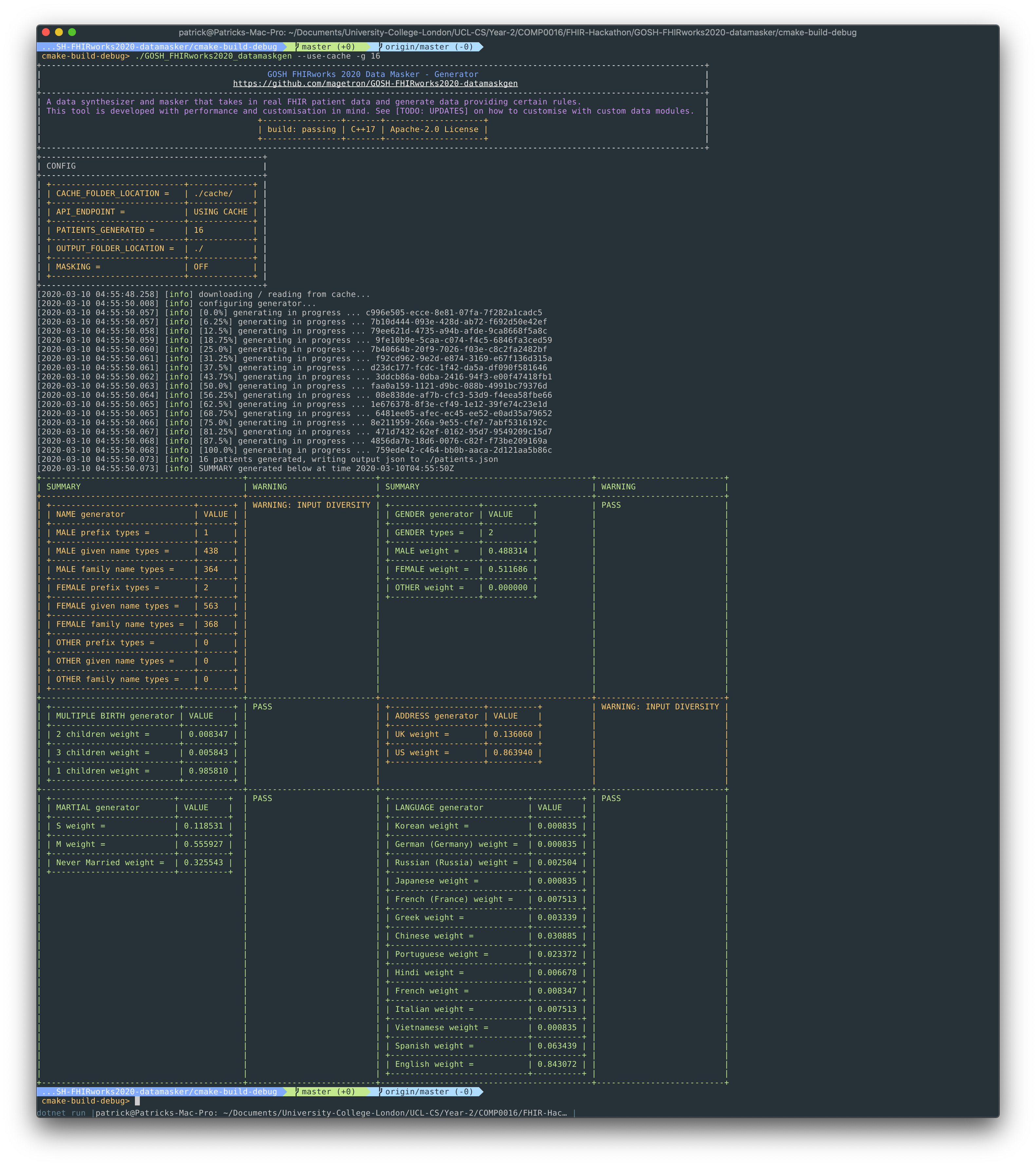
A data synthesizer and masker that takes in real FHIR patient data and generate data providing certain rules.
This tool is developed with performance and customisation in mind. See below on how to customise with custom data modules.
mkdir build
cd build
cmake ..To compile and use under current folder,
makeOr to install in system (POSIX system only),
make installNote : To be used with Patients API endpoints, official version from GOSH, or unofficial ones here from @henryz00, or simply a gist - Github, whichever you chose, please specify when calling the application.
--quietforSILENTMode--use-cachefor loading cachedpatients.jsonfile at a custom location, to be used with--cache-loc--api ${YOUR_CUSTOM_API}--cache-loc ${CACHE_FILE_LOCATION}-g ${NO OF PATIENTS TO BE GENERATED}-o ${OUTPUT_FILE_LOCATION}-hor--helpfor showing the help message
./GOSH_FHIRworks2020_datamaskgen --api https://localhost:5001/api/Patient -g 100 -o ./Build with performance in mind, Data MaskGen utilises system resources much better than the industrial standard synthea and therefore archives much better running time with similar results.
| Patient Amount | Data MaskGen | Synthea |
|---|---|---|
| 1 | 98% cpu, 1.836s | 10% cpu, 13.067s |
| 50 | 99% cpu, 1.863s | 9% cpu, 15.817s |
| 100 | 99% cpu, 1.917s | 6% cpu, 23.035s |
| 500 | 99% cpu, 2.258s | 5% cpu, 30.726s |
| 1000 | 99% cpu, 2.601s | 4% cpu, 41.757s |
Customise any of the generator in the src/generator folder, or add your own class with suffix _generator.
Your customised class will be required to feed in a const reference to original patients list vector, and then generate required information based own your customised algorithm.
Specify your output of custom data set in jsonify() function in src/patient/patient.hh, and sit back to see the new data getting generated.