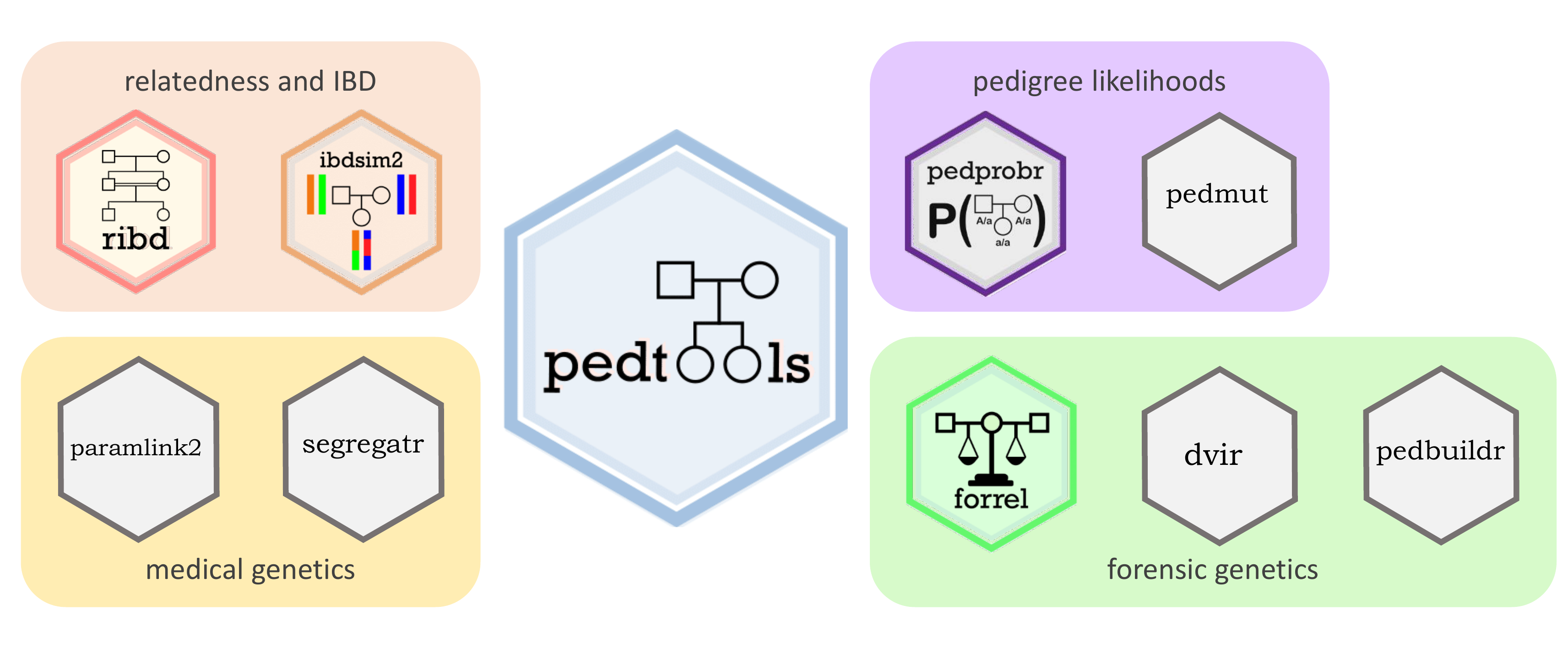
The pedsuite is a collection of R packages for pedigree analysis, covering a variety of applications across several fields. The packages are designed to work harmoniously together, often allowing complex tasks to be solved in a few lines of code.
At the centre of the pedsuite we find the package pedtools, on which all the others depend. In addition, several packages contain basic features often needed in many types of analysis. These are the core packages:
- pedtools: Creating and working with pedigrees and marker data
- forrel: Forensic pedigree analysis and relatedness analysis
- pedprobr: Marker probabilities and pedigree likelihoods
- ribd: Computation of pedigree-based relatedness coefficients
- verbalisr: Verbal descriptions of pedigree relationships
The following packages are devoted to special applications:
- ibdsim2: Simulation of identity-by-descent sharing by family members
- dvir: Disaster victim identification
- paramlink2: Parametric linkage analysis
- pedbuildr: Pedigree reconstruction
- pedFamilias: Import and export .fam files used by the Familias software
- pedmut: Mutation models for pedigree likelihood computations
- segregatr: Segregation analysis for clinical variant interpretation
These packages have been superseded by other packages and are no longer compatible with the rest of the pedsuite. They are maintained only for legacy purposes and should not be used in new projects.
- paramlink: This package marked the birth of the pedsuite. Originally intended for parametric linkage analysis, it also contained the seeds of the current packages pedtools, pedprobr, ribd and forrel.
- IBDsim: This is replaced by ibdsim2. (Regrettably, the name ‘IBDsim’ is very similar to that of the unrelated (non-R) software ‘IBDSim’ (Leblois, Estoup & Rousset).)
- kinship2: Pedigree plotting in the pedsuite depends on kinship2 (Sinnwell, Therneau & Schaid) for pedigree alignment calculations.
Several Shiny apps based on the pedsuite have been developed. Read more about them here:
- QuickPed: An online pedigree creator
- KLINK: Kinship analysis with linked markers
- ibdsim2-shiny: IBD distributions in pedigrees
- LinkageLab: Playing with linked markers
- ibdClassifier: Classifying relationships based on their shared IBD segments