Code for the paper "CitationIE: Leveraging the Citation Graph for Scientific Information Extraction" at ACL 2021.
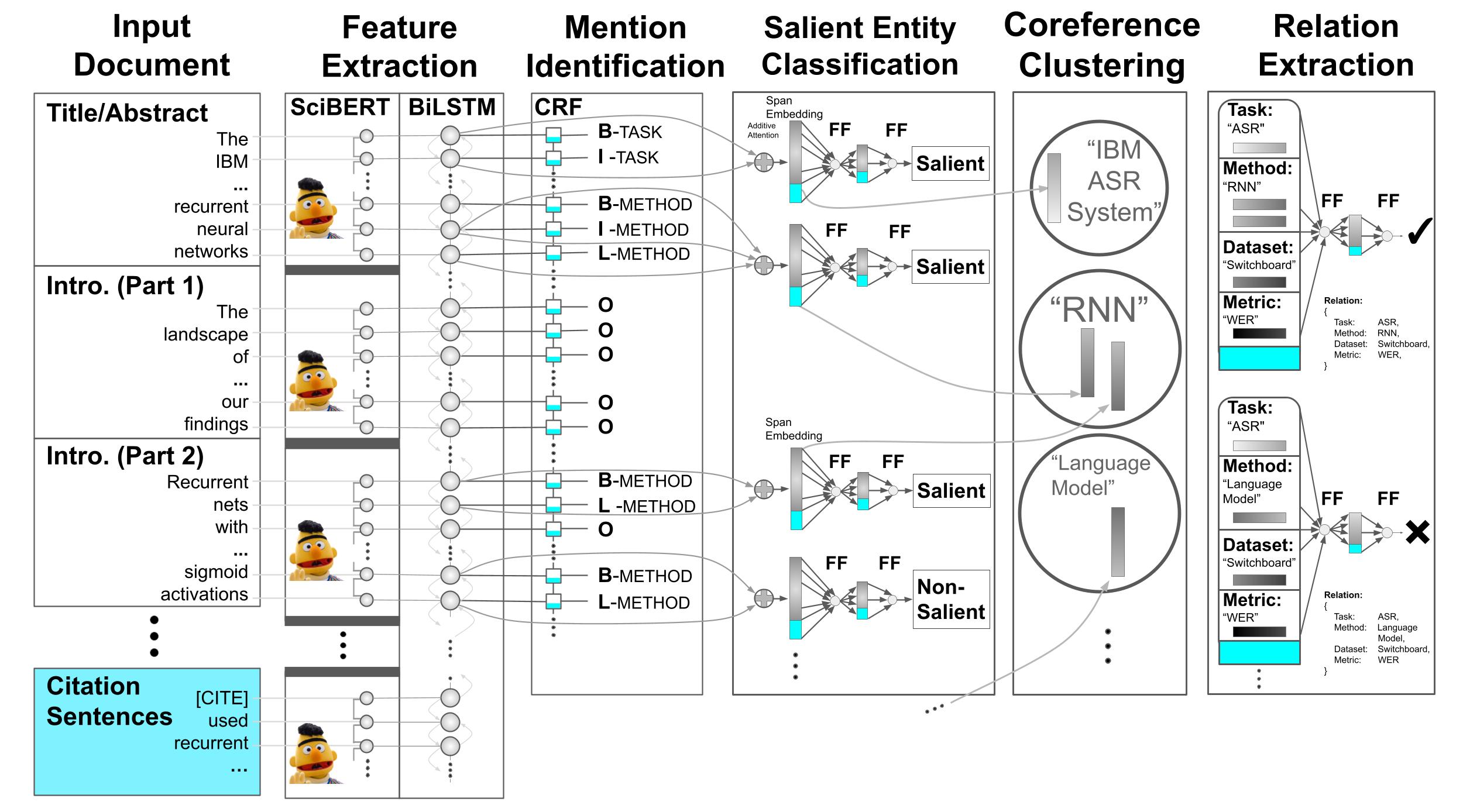 This repository serves two purposes:
This repository serves two purposes:
- Provides tools for joining the SciREX dataset with the S2ORC citation graph.
- Provides model training functionality to build models on the joined data from 1)
We provide three utilities that may be useful independent of any of the code in this repository:
- A mapping of SciREX document IDs to S2ORC IDs (which are also all valid SemanticScholar IDs) is
scirex_to_s2orc_id_mapping.json - Pretrained graph embeddings for each document in SciREX is in
SciREX/graph_embeddings/ - A version of the SciREX dataset where each document is augmented with citation sentences can be found at
SciREX/scirex_dataset/data_with_citances/
Beyond these utilities, we also have a set of tools available for constructing citation graphs for the purpose of information extraction research. If you want to run any of our software, you must:
- Download and untar the SciREX dataset
- Request access to the S2ORC dataset, managed by the Allen Institute for AI
- Update
metadata_downloads.shandfull_data_downloads.shwith the scripts given to you by the AllenAI team (these scripts contain API keys that we scrape and use in this library)
After this, run python join_scirex_and_s2orc.py. This script will run for at least 5 hours, require at least 3 GB of disk space, and download over a hundred GB of S2ORC data over the internet (though this data is deleted as soon as it is used).
Once this is done, you can do:
from join_scirex_and_s2orc import get_scirex_to_s2orc_mappings, get_citation_graph, S2OrcEntry, S2Metadata
scirex_to_s2orc_mapping = get_scirex_to_s2orc_mappings() # Find S2ORC/S2 ids for a given SciREX document
citation_graph = get_citation_graph(radius=2) # Construct a citation graph of all documents within 2 hops of a document in SciREX
Giving an adjacency-list representation of the citation graph surrounding documents in the SciREX dataset.
To install all dependencies for model training, please follow the instructions in the SciREX repository. In particular, you will need to untar the SciREX dataset, and download a trained copy of SciBERT to your machine. All commands listed here assume you are at the root of the SciREX directory (which is a submodule of this repository).
We provide scripts for training 4 kinds of models:
- End-to-End Information Extraction (
scirex/commands/train_scirex_model.sh) - Mention Identification Only (
scirex/commands/train_ner_only.sh) - Salient Entity Classification Only (
scirex/commands/train_salient_classification_only.sh) - Relation Extraction Only (
scirex/commands/train_relations_only.sh)
For training baseline models for each, run the following commands from the SciREX/ directory, on a CUDA-enabled machine:
export BERT_BASE_FOLDER=<PATH_TO_SCIBERT>
CUDA_DEVICE=0 bash <TRAINING_SCRIPT> main
In order to train citation graph-enhanced models for any of the above, set the following environment variables before running the above commands:
export use_citation_graph_embeddings=true
export citation_embedding_file=graph_embeddings/embeddings.npy
export doc_to_idx_mapping_file=graph_embeddings/scirex_docids.json
In order to train citance-enhanced models, do export DATA_BASE_PATH=scirex_dataset/data_with_citances before running your desired training command.
Depending on the type of model you trained, use one of the following evaluation scripts
scirex/commands/predict_scirex_model.shscirex/commands/predict_ner_only_gold.shscirex/commands/predict_salient_only_gold.shscirex/commands/predict_salient_only_gold.sh
And then, run:
export BERT_BASE_FOLDER=scibert_scivocab_uncased
export PYTHONPATH=$PYTHONPATH:$(pwd)
export scirex_archive=<PATH TO TRAINED MODEL>>
export scirex_coreference_archive=<PATH TO TRAINED COREFERENCE MODEL
export cuda_device=2
export test_output_folder=<PREDICTION OUTPUTS DIRECTORY>
bash scirex/commands/predict_salient_only_gold.sh
We also have written a number of additional evaluation scripts for the results in our paper, which are all in scirex/evaluation_scripts:
Bootstrap Evaluation Scripts
relation_bootstrap_comparison.py
relation_bootstrap_comparison_multi_model.py
salient_bootstrap_comparison.py
salient_bootstrap_comparison_multi_model.py
Bucketing and Visualization Scripts relation_bootstrap_comparison_bucketing_on_cluster_distance.py relation_bootstrap_comparison_bucketing_on_graph_degree.py relation_bootstrap_comparison_bucketing_on_graph_degree_multi_model.py
salient_bootstrap_comparison_bucketing_on_graph_degree.py
@inproceedings{CitationIE,
title = {CitationIE: Leveraging the Citation Graph for Scientific Information Extraction},
author = {Vijay Viswanathan and Graham Neubig and Pengfei Liu},
booktitle = {Joint Conference of the 59th Annual Meeting of the Association for Computational Linguistics and the 11th International Joint Conference on Natural Language Processing (ACL-IJCNLP)},
address = {Virtual},
month = {August},
year = {2021}
}