-
Authors: Mahsa Sarafrazi, Rowan Sivanandam, Shiva Jena, and Vanessa Yuen
-
Contributors: Instructors, and Teaching Assistants of the course DSCI 522 of UBC Master of Data Science Vancouver program at the University of British Columbia, Vancouver
Analytical project prepared as part of the course DSCI 522 of the Master of Data Science 2021-22 program at University of British Columbia, Vancouver.
This analytical project is an attempt at creating a machine learning linear regression model to predict a continuous variable, weight of giant pumpkins based on features such as type of pumpkin, place of cultivation (country, city, state province, GPC site), seed origins, pollinator father, etc.
The data set used in this project is originally from BigPumpkins.com. The Great Pumpkin Commonwealth’s (GPC) mission promotes the hobby of cultivating giant pumpkins throughout the world through standards and regulations to ensure quality of fruit, fairness of competition, recognition of achievement, fellowship and education for all participating growers and weigh-off sites. (Read more from (“The Great Pumpkin Commonwealth” 2021) here and (“Bigpumpkins.com: A Giant Pumpkin Growing Community,” n.d.) here).
The dataset is a public domain resource which pertains to the attributes of giant pumpkins grown in around 20 countries across the world in different regions. The raw data which was used in this project for the analysis can be found here.
Given certain features of the seeds, place of cultivation, features of the parent pumpkin etc, what will be estimated weight of the giant pumpkin after harvesting?
To answer this predictive question, we need to first answer some underlying data related questions such as:
-
Is there a relationship between features of the seeds, place of cultivation, and the weight of the pumpkins?
-
How are the weights distributed across regions?
The majority of the raw data comprises of features of the character type
where some of the features such as id contain important information
such as the pumpkin type. Therefore, in order to proceed, an initial
data cleaning, preparation and pre-processing is required to make the
features ready for training purposes. However, since the analysis is an
attempt to answer a machine learning prediction problem, the dataset is
first split into a 70/30 split for the training and test sets
respectively along with random seeding for reproducibility. Brief
details of the steps are mentioned below for an outline:
-
Train Test Split
Splitting the dataset into train and test splits along with random seed for reproducibility.The desired outputs are a 70/30 split of training and test data.
-
Exploratory Data Analysis (EDA):
Data cleaning and preparation is required for making features ready for the machine learning regression model. An analysis of the trends and correlation with actual pumpkin weights and various features will be used using Python (Van Rossum and Drake 2009) and Altair (VanderPlas et al. 2018) as visualisation tools.
On initial observations, the data seems to be mostly from the US. The distribution of the GPC sites, city and state/province are more evenly distributed. We consider these columns as good features to be used. Plots of the mean weight of giant pumpkins against different features (ott, country, city, state, gpc site) also suggest these features relates to the target (weight).
The processed data sets in form of .csv files.
-
Predictive Modelling
The Ridge Linear Regression model will be used as pumpkin weight is a continuous, quantitative, numerical variable. The model is planned to be trained and tested using Scikit Learn (Pedregosa et al. 2011) packages.
There are few numerical features and more categorical features. For numerical features, SimpleImputer and StandardScaler will be used during the preprocessing stage where as for categorical features, One-Hot Encoding and SimpleImputer for will ready the data for analysis.
Using column transformers and pipe operators, cross-validation will be performed for hyperparameter optimization of sklearn’s LinearRegression model using GridSearchCV.
After optimising the hyperparameters, the model will be fit on the training set and evaluation to be done on the test set. In the initial stages, the R-squared score will be the underlying metrics used to assess our model.
Results of the analysis can be found here .
For replicating the analysis and usage, there are two ways:
1. Using Docker
note - the instructions in this section also depends on running this in a unix shell (e.g., terminal or Git Bash).
-
Install Docker
-
Download/clone this repository in the path you have selected.
-
Use the command line to navigate to the root of this downloaded/cloned repo
-
Type the following:
docker run --rm -v "/$(pwd):/home/rstudio/pumpkin" imtvwy/giant_pumpkin_weight_prediction make -C home/rstudio/pumpkin all -
To reset/undo the analysis, type the following commands:
docker run --rm -v "/$(pwd):/home/rstudio/pumpkin" imtvwy/giant_pumpkin_weight_prediction make -C home/rstudio/pumpkin clean
-
Download/clone this repository and using the command line, navigate to the root of this project.
-
install the project environment(
environment.yaml) -
To run the analysis, type the following commands:
make all -
To reset/undo the analysis, type the following commands:
make clean
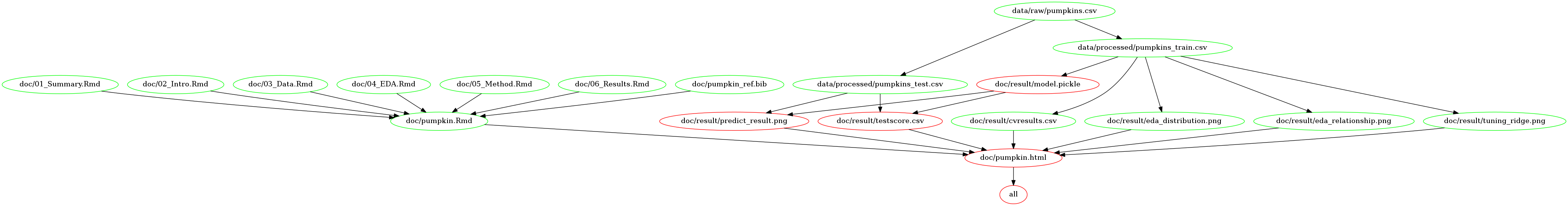
Below image can demonstrate the dependency diagram for making the report
by Makefile
The dependencies for this project can be found in
environment.yaml.
The yaml file needs to be run to create the environment required for running the analysis. If conda is installed, the following command can be run at the terminal/ command line from the root directory of this project folder (“…/Giant_Pumpkins_Weight_Prediction/”) to install the dependencies:
conda activate pumpkin
After having finished the work with our project, the following command can be run to deactivate the environment in the terminal/command line:
conda deactivate
The list of dependencies are given below which are tentative and may change as per updates in the environment file:
-
Python & Python libraries:
altair_saver==0.5.0=py_0requests[version='>=2.24.0']pandas[version='>=1.3.*']vega_datasets=0.9.0ipykernel=6.5.1altair==4.1.0=py_1graphviz=2.49.3python-graphviz=0.19scikit-learn[version='>=1.0']altair_data_server==0.4.1=py_0pandoc=2.16.2altair_viewer==0.4.0=pyhd8ed1ab_0matplotlib[version='>=3.2.2']docopt=0.6.2 -
GNU make 4.2.1
For generating the plots in the report, install the following packages with this command in the environment created:
conda activate pumpkin
npm install -g vega vega-cli vega-lite canvas
If the above is already installed, the file may already exist and therefore, it may throw an error asking to force install, which may be ignored.
“Bigpumpkins.com: A Giant Pumpkin Growing Community.” n.d. BigPumpkins.com: A Giant Pumpkin Growing Community. http://www.bigpumpkins.com/.
Pedregosa, F., G. Varoquaux, A. Gramfort, V. Michel, B. Thirion, O. Grisel, M. Blondel, et al. 2011. “Scikit-Learn: Machine Learning in Python.” Journal of Machine Learning Research 12: 2825–30.
“The Great Pumpkin Commonwealth.” 2021. The Great Pumpkin Commonwealth | Our Organization Promotes Encourages and Promotes the Participation in Growing Giant Fruits and Vegetables. https://gpc1.org/.
Van Rossum, Guido, and Fred L. Drake. 2009. Python 3 Reference Manual. Scotts Valley, CA: CreateSpace.
VanderPlas, Jacob, Brian Granger, Jeffrey Heer, Dominik Moritz, Kanit Wongsuphasawat, Arvind Satyanarayan, Eitan Lees, Ilia Timofeev, Ben Welsh, and Scott Sievert. 2018. “Altair: Interactive Statistical Visualizations for Python.” Journal of Open Source Software 3 (32): 1057.