Contributors:
- Dongxiao Li
- Kyle Maj
- Mahmoodur Rahman
Data analysis project executed by Group 4.0 of 6th Cohort (2021-22) for DSCI 522 (Data Science workflows); a course in the Master of Data Science program at the University of British Columbia.
A project proposal is discussed here.
Mushrooms are species of fungus, of which some can be eaten with meaty texture and few types are toxic 1. Annually, a significant number of people die from ingesting poisonous mushrooms 2,3. The BC Centre for Disease Control (BCCDC) received 200 calls relating to mushroom poisoning in 2018 4. Thus, It is critical to recognize a mushroom of poisonous species by observing it's appearance. By appearance, primarily it refers to certain physical traits. A model recognizing mushroom toxicity by taking these physical traits into account can be effective at preventing mushroom toxicity 5. Recent methods on classifying mushroom falls into three groups, chemical determination, animal experimentation, fungal classification and folk experience 6. These methods are not perfect and there is room for improvement 7. Mankind has been identifying toxic mushrooms by observing morphology, smell and distinct features 8. These intuitive-based methods are less reliable, and often lead to fatal incidents. However, relying on these experiences and intuitions, machine-learning models can be tried and tested. In this era of fourth Industrial revolution, artificial intelligence is playing a major role through deployment of machine-learning and deep learning model 9. This also made it's way to detecting poisonous muashroom also. Chaoqun and colleagues developed an android-based application, which detects toxic mushrooms through machine-learning models 10. To further improve classification, decision fusion method has been used, by stacking algorythms 11. Shuaichang and colleagues used image-based models for poisonous mushroom detection 12.
The purpose of this project is to create a model that can be used to identify poisonous mushrooms with a greater degree of accuracy than human intuition. We selected our model through a combination of exploratory data analysis on our data set and a comparison of cross-validation training scores between several candidate models. Once our model was selected we applied standard optimization and tuning processes before assessing its performance on out test data. Our chosen scoring metrics indicate that this model performs extremely well on test data. While these results are encouraging, we feel that it would be beneficial to do a second pass of this project in the future in order to apply more rigorous optimization tools, calibrate for type I errors, and examine possible sources of overfitting.
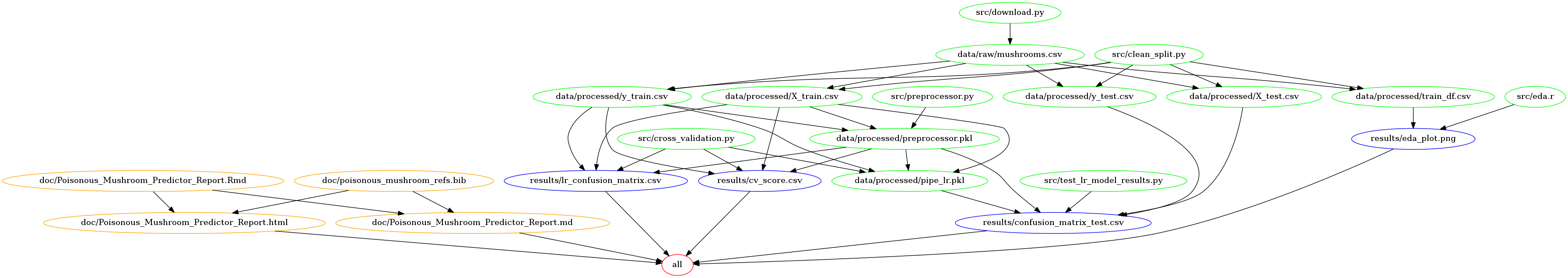
Our project summary workflow
Data used in this project is from UCI machine learning repository, provided by Jeff Schlimmer of the Audubon Society Field Guide to North American Mushrooms 13. The dataset has 8124 examples and 22 features with 2 possible target classes (edible and poisonous). The features are: cap shape, cap surface, cap color, bruises, odor, gill spacing, gill attachment, gill size, gill color, stalk shape, stalk root, stalk surface above ring, stalk surface below ring, stalk color above ring, stalk color below ring, veil type, veil color, ring number, ring type, spore print color, population, and habitat.
The final report can be found here.
There are two suggested ways to run this analysis:
- Using Docker note - the instructions in this section also depends on running this in a unix shell (e.g., terminal or Git Bash)
To replicate the analysis, install Docker. Then clone this GitHub repository and run the following command at the command line/terminal from the root directory of this project:
docker run --rm -v "/$(pwd):/home/rstudio/poisonous_mushroom_predictor" dol23/docker_mushroom make -C /home/rstudio/poisonous_mushroom_predictor all
To reset the repo to a clean state, with no intermediate or results files, run the following command at the command line/terminal from the root directory of this project:
docker run --rm -v "/$(pwd):/home/rstudio/poisonous_mushroom_predictor" dol23/docker_mushroom make -C /home/rstudio/poisonous_mushroom_predictor clean
If the above commands do not work on windows please use the following.
Run the analysis:
docker run --rm -v "/$(pwd):/home/rstudio/Poisonous_Mushroom_Predictor" dol23/docker_mushroom make -C //home//rstudio//Poisonous_Mushroom_Predictor all
Clear intermediate and results files:
docker run --rm -v "/$(pwd):/home/rstudio/Poisonous_Mushroom_Predictor" dol23/docker_mushroom make -C //home//rstudio//Poisonous_Mushroom_Predictor clean
- Without using Docker
To replicate the analysis, clone this GitHub repository, install and activate the environment file listed below, and run the following command at the command line/terminal from the root directory of this project:
Install and activate the environment file:
conda env create -f env-mushroom.yaml
conda activate mushroom
Then navigate to the directory of our project repository, run the command below in the terminal:
make all
To reset the repo to a clean state, with no intermediate or results files that are needed to generate the final report, run the following command at the command line/terminal from the root directory of this project:
make clean
Original commands for scripts needed(they are all included in the Makefile):
# download mushrooms data set to directory
python src/download.py --url="https://raw.githubusercontent.com/kanchitank/Mushroom-Classification/master/mushrooms.csv" --out_file=data/raw/mushrooms.csv
# clean and split the raw data into test and train parts and write to assigned directory
python src/clean_split.py --input_csv="data/raw/mushrooms.csv" --out_dir="data/processed/"
# create a preprocessor to transform the train data that can be passed to the next script
python src/preprocessor.py --X_train_input='data/processed/X_train.csv' --y_train_input='data/processed/X_train.csv' --out_pkl='data/processed/preprocessor.pkl'
# cross validation and confusion matrix on train data
python src/cross_validation.py --X_train_input='data/processed/X_train.csv' --y_train_input='data/processed/y_train.csv' --out_cv='results/cv_score.csv' --out_matrix='results/lr_confusion_matrix.csv' --out_pkl='data/processed/pipe_lr.pkl'
# test the model and output results(confusion matrix)
python src/test_lr_model_results.py --X_test_input='data/processed/X_test.csv' --y_test_input='data/processed/y_test.csv' --out_matrix_test='results/confusion_matrix_test.csv'
To set up the necessary packages for running the data analysis materials from poisonous mushroom prediction, download the environment file from the repo to your computer (hit "Raw" and the Ctrl + s to save it, or copy paste the content). Then create a Python virtual environment by using conda with the environment file you just downloaded:
conda env create -f env-mushroom.yaml
conda activate mushroom
More information about dependencies can be referred to our Dockerfile.
The Poisonous Mushroom Prediction materials here are licensed under the Creative Commons Attribution 2.5 Canada License (CC BY 2.5 CA). If re-using/re-mixing please provide attribution and link to this webpage.
Footnotes
-
J. H. Tegzes and B. Puschner, ““Toxic mushrooms,” the veterinary clinics of north America,” Veterinary Clinics of North America: Small Animal Practice, vol. 32, no. 2, pp. 397–407, 2002. ↩
-
C. Lei, W. Tangkanakul, and L. Lu, “Mushroom poisoning surveillance analysis,” OSIR Journal, vol. 1, no. 1, pp. 8–11, 2006. ↩
-
J. White, S. A. Weinstein, L. De Haro et al., “Mushroom poisoning: a proposed new clinical classification,” Toxicon, vol. 157, pp. 53–65, 2019. ↩
-
http://www.bccdc.ca/about/news-stories/stories/mushroom-poisonings-on-the-rise-in-british-columbia ↩
-
J. H. Diaz, “Evolving global epidemiology, syndromic classification, general management, and prevention of unknown mushroom poisonings,” Critical Care Medicine, vol. 33, no. 2, pp. 419–426, 2005. ↩
-
T. Fukuwatari, E. Sugimoto, K. Yokoyama, and K. Shibata, “Establishment of animal model for elucidating the mechanism of intoxication by the poisonous mushroom Clitocybe acromelalga,” Journal of the Food Hygienic Society of Japan (Shokuhin Eiseigaku Zasshi), vol. 42, no. 3, pp. 185–189, 2001. ↩
-
M. Lu, “Present status and future prospects of the mushroom industry in China,” Acta Edulis Fungi, vol. 13, no. 1, pp. 1–5, 2006. ↩
-
K. Tanaka, S. Miyasaka, and T. Inoue, “Histopathological effects of illudin S, a toxic substance of poisonous mushroom, in rat,” Human & Experimental Toxicology, vol. 15, no. 4, pp. 289–293, 1996. ↩
-
W. A. Reynolds and F. H. Lowe, “Mushrooms and a toxic reaction to alcohol,” New England Journal of Medicine, vol. 272, no. 12, pp. 630-631, 1965. ↩
-
Z. Chaoqun, Recognition and Research of Poisonous Mushroom Based on Machine Learning, Taigu: Shanxi Agricultural University, Jinzhong, China, 2019. ↩
-
Y. Zhifeng, Application of Multi-Classifier Fusion Based on Stacking Algorithm in Identification of Poisonous Mushrooms, Taigu: Shanxi Agricultural University, Jinzhong, China, 2019. ↩
-
F. Shuaichang, Y. Xiaomei, and L. Jian, “Toadstool image recognition based on deep residual network and transfer learning,” Journal of Transduction Technology, vol. 33, no. 1, pp. 74–83, 2020. ↩