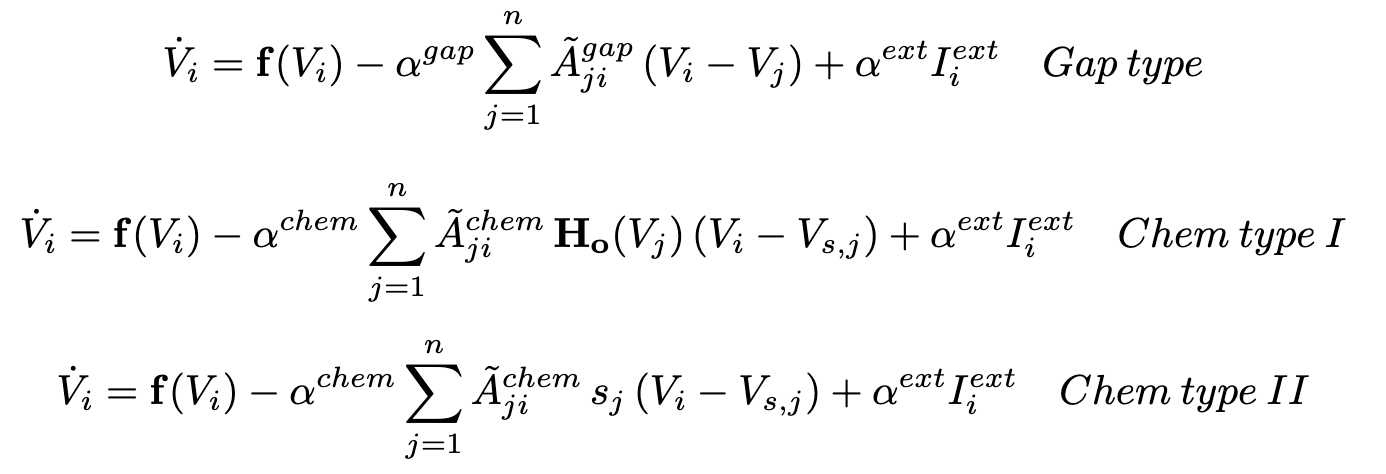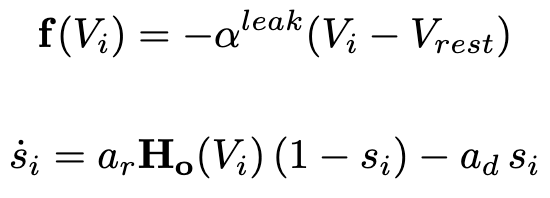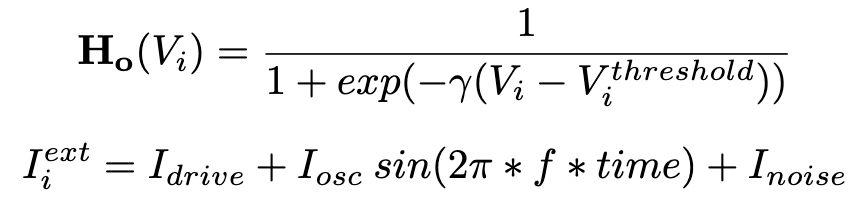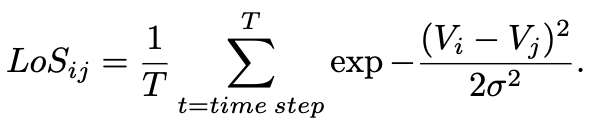Ready to install and run MatLab app (CelegansLocomotion.mlappinstall) simulates the forward and backward locomotive neural network dynamics with multiple control variables.
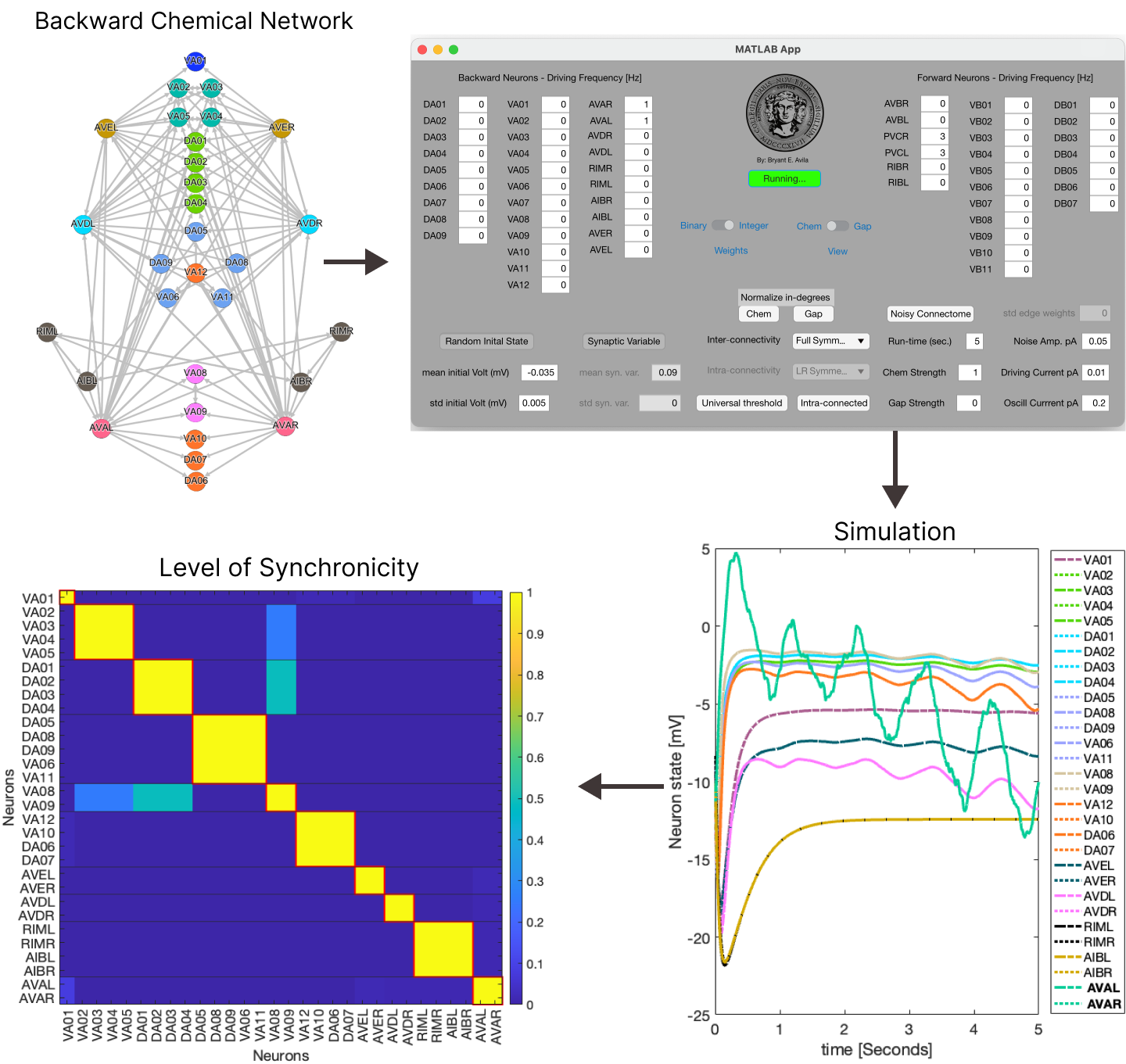 The user can specify the type of networks used (electron micrograph connectivity produced by Varshney [1], left-right symmetrized, fully symmetrized [2]) for the forward+backward network system as well as the type for the inter-connectivity between these two. The inter-connectivity interactions are considered inhibitory while intra-connectivity interactions are considered exitatory.
The user can specify the type of networks used (electron micrograph connectivity produced by Varshney [1], left-right symmetrized, fully symmetrized [2]) for the forward+backward network system as well as the type for the inter-connectivity between these two. The inter-connectivity interactions are considered inhibitory while intra-connectivity interactions are considered exitatory.
The type of ordinary differential equation models that can be used for neuron interactions are captured in the equtions below. The strength of the gap junctions and chemical synapses interactions can be specified and allow a network to only have either of these interaction or any combination of strength between these two with each strength being expressed between the values of 0 to 1.
Addiontionaly it is possible to specify the type of weights used for the connectivity (binary or integer) and the ability to randomly alter these weights, normalize the in-degree of each neuron for both types of interactions, the initial distribution of voltages and synaptic variables, type of model used for the chemical synapses interactions and more controlls. Fianlly one is able to specify which neurons receive a static input current, the amplitude and frequency of an undulatory input and a Gaussian random walk input. Dynamics are evolved using a Runge-Kutta-4th method. Within the folder "network" are all the files needed to generate all the networks in our manuscript. All of these are integer weighted, binarization happens within the code. After the simulation has concluded the app also calculates the level of synchronicity (LoS) as per the equation below for the pair of neurons for each of the two networks for the last second of simulation time. Other measurements are also accesible (Phase Locking Value, Phase Differenece and Standard Deviation Difference).Toolboxes that are needed:
Statistic and Machine Learning
Fuzzy Logic
[1] Lav R Varshney, Beth L Chen, Eric Paniagua, David H Hall, and Dmitri B Chklovskii. Structural properties of the caenorhabditis elegans neuronal network. PLoS computational biology, 7(2):e1001066, 2011
[2] Flaviano Morone and Hernan A Makse. Symmetry group factorization reveals the structure-function relation in the neural connectome of caenorhabditis elegans. Nature communications, 10(1):1–13, 2019
[3] Bryant Avila, Hernan Makse et. al. Fibration symmetries and cluster synchronization in the Caenorhabditis elegans connectome (Under review)
as also seen at @kryogenica
For any questions please contact: bryant.avila.physics@gmail.com or hmakse@ccny.cuny.edu
This project is licensed under the MIT License - see the LICENSE.md file for details