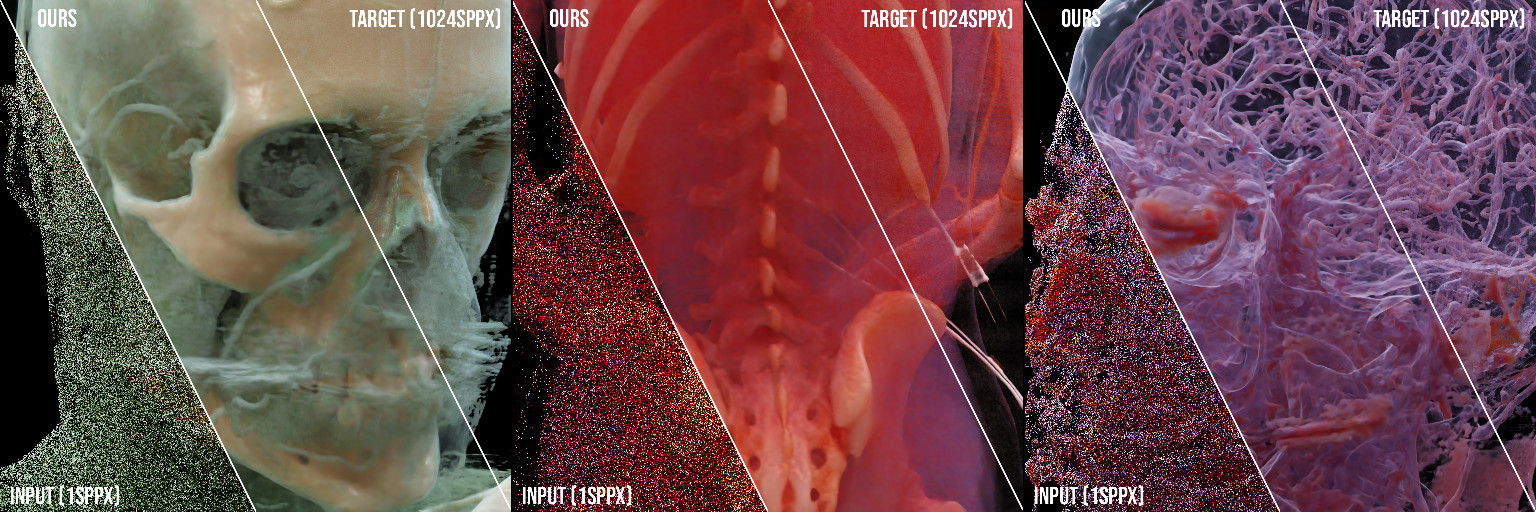
Auxiliary source material for our HPG'20 paper "Neural Denoising for Path Tracing of Medical Volumetric Data": https://doi.org/10.1145/3406181
We transfer machine learning techniques previously applied to denoising surface-only Monte Carlo renderings to path-traced visualizations of medical volumetric data. In the domain of medical imaging, path-traced videos turned out to be an efficient means to visualize and understand internal structures, in particular for less experienced viewers such as students or patients. However, the computational demands for the rendering of high-quality path-traced videos are very high due to the large number of samples necessary for each pixel. To accelerate the process, we present a learning-based technique for denoising path-traced videos of volumetric data by increasing the sample count per pixel; both through spatial (integrating neighboring samples) and temporal filtering (reusing samples over time). Our approach uses a set of additional features and a loss function both specifically designed for the volumetric case. Furthermore, we present a novel network architecture tailored for our purpose, and introduce reprojection of samples to improve temporal stability and reuse samples over frames. As a result, we achieve good image quality even from severely undersampled input images.
Setup a virtual python environment:
virtualenv -p python3 env
source env/bin/activate
pip3 install -r requirements.txt
Single autoencoder architecture:
python src/train.py /path/to/data/noisy /path/to/data/clean /path/to/data/validation/noisy /path/to/data/validation/clean
Dual autoencoder architecture:
python src/train.py /path/to/data/noisy /path/to/data/clean /path/to/data/validation/noisy /path/to/data/validation/clean -t dual
GAN-based dual autoencoder architecture:
python src/train_relgan.py /path/to/data/noisy /path/to/data/clean /path/to/data/validation/noisy /path/to/data/validation/clean --big
Dual autoencoder architecture with temporal reprojection:
python src/train_reproj.py /path/to/data/noisy /path/to/data/clean /path/to/data/validation/noisy /path/to/data/validation/clean --feature_select
Additional command line arguments are listed in the beginning of each source file.
python src/denoise.py /path/to/model.pt /path/to/data/noisy /path/to/data/clean
Additional command line arguments are listed in the beginning of the source file.
Pretrained models for all configurations are available at models/*.pt. To load a model into pytorch or access the weights:
cd src/
python3
>>> import torch
>>> import models
>>> torch.load('../models/single_color_only.pt').state_dict()
For each stored frame under /path/to/data/noisy the following data is expected:
- <Frame>.hdr: input color data
- <Frame>_pos.hdr: primary scatter positions
- <Frame>_norm1.hdr: primary normals
- <Frame>_norm2.hdr: color after first surface approximation
- <Frame>_alb1.hdr: primary albedo
- <Frame>_alb2.hdr: secondary albedo
- <Frame>_vol1.hdr: primary volumetrics
- <Frame>_vol2.hdr: secondary volumetrics
For each stored frame under /path/to/data/clean the following data is expected:
- <Frame>.hdr: target color data
For temporal reprojection, each noisy frame is additionally expected to provide:
- <Frame>_pos.dat: binary high-precision primary scatter positions
- <Frame>_matrices.txt: three 4x4 floating point matrices (model, view, proj), i.e. 12 rows with 4 IEE plaintext floats each
File names are recursively globbed and then sorted alphanumerically, thus file names do not need to match exactly between noisy and clean data, just required to sort into the same order.
Unfortunately, due to their medical nature and data protection laws, it is not possible for us to publish the training datasets. We plan to extend this repository by a freely available dataset.