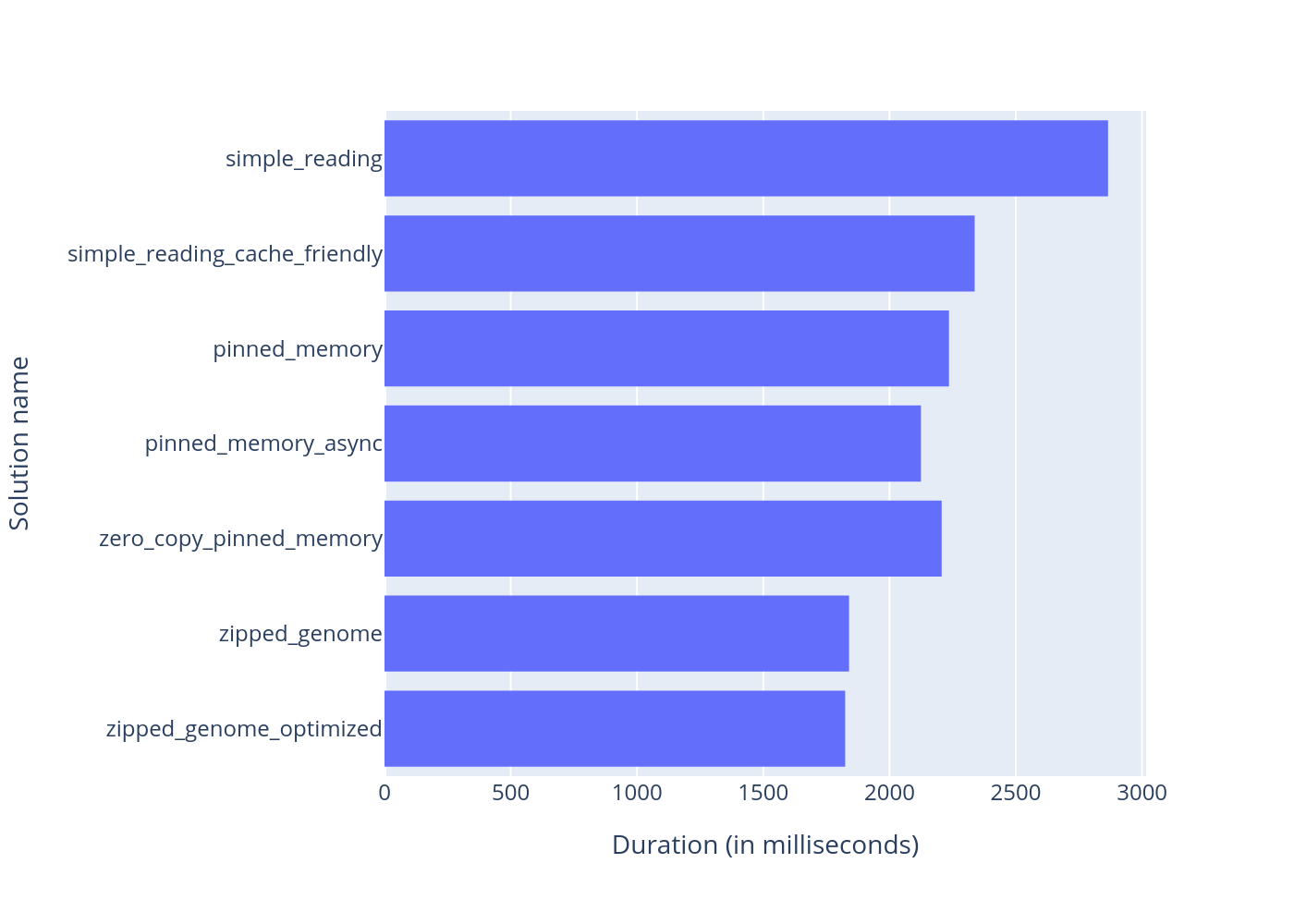
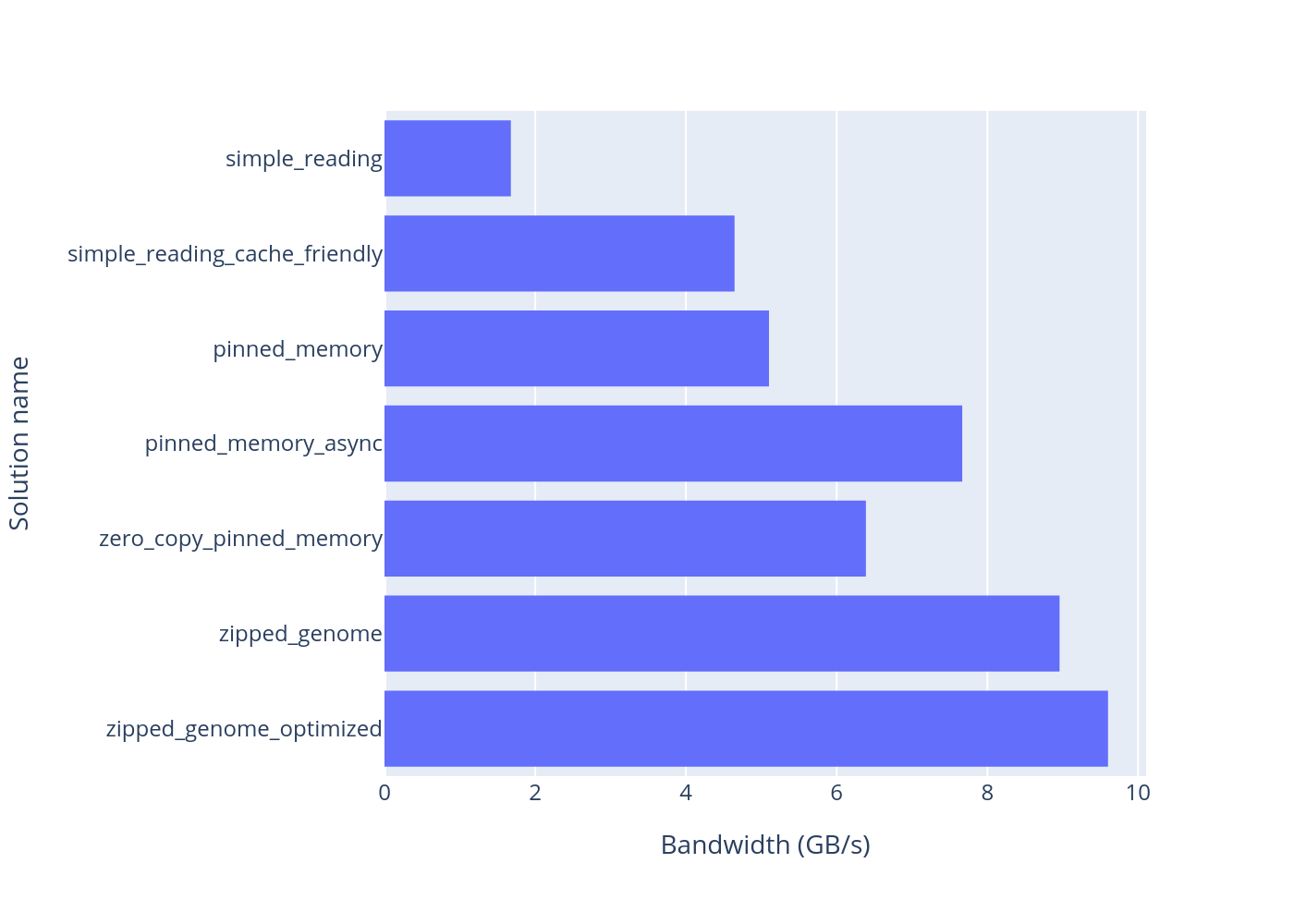
This is a CUDA project that is designed to compare different approaches for reading data from SSD to GPU for future analyzing of that data. For testing data I chose to work with genomes and all the results wil be shown for short fake 100 MB long genomes.
For generating fake 100 MB long genomes there is small project written in data directory.
Usage:
cd data_generator
cmake -B build -D CMAKE_BUILD_TYPE=Release
cmake --build build
./data_generator <GENOMES_NUM> <THREADS_NUM>Usage of data_generator:
Usage: ./data_generator <GENOMES_NUM> <THREADS_NUM>
<GENOMES_NUM> - number of genomes to generate
<THREADS_NUM> - number of threads to use for generation
Note: The performance of analyzing 12 randomly generated genomes are tested solutions is tested on NVIDIA GeForce GTX 1650.
Comparison of performance of different CUDA memory management algorithms.
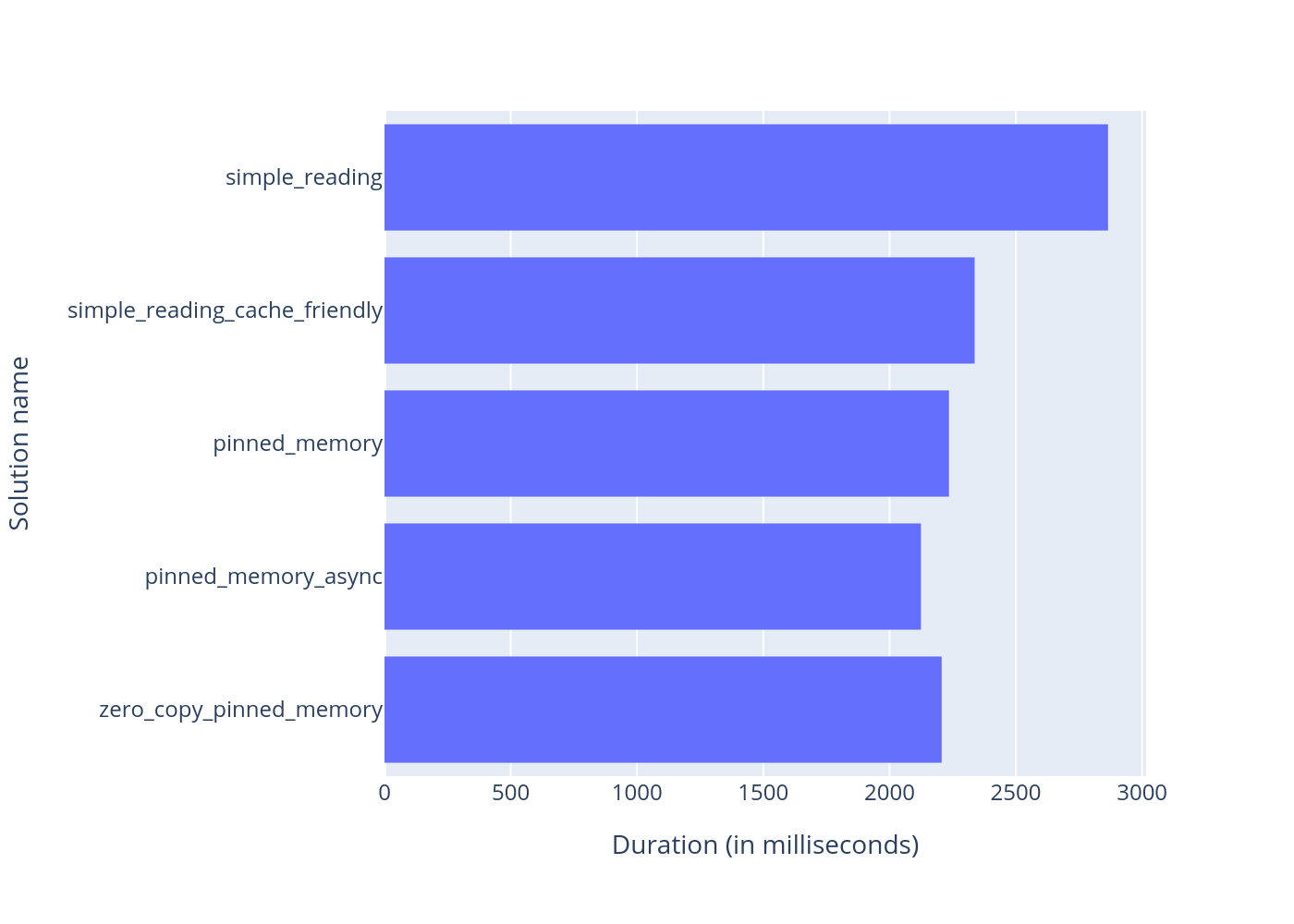
To squeeze the performance even more, we can add additional software optimization, such as genome compressing and micro optimizations:
Comparison of GPU bandwidth (in GB per second) for each solution:
Build all solutions:
cmake -B build -D CMAKE_BUILD_TYPE=Release
cmake --build buildRun solution:
cd ./solutions/<SOLUTION_NAME>/ && \
../../build/solutions/<SOLUTION_NAME>/<SOLUTION_NAME> && \
cd ../..For instance:
cd ./solutions/part_1_simple_reading/ && \
../../build/solutions/part_1_simple_reading/part_1_simple_reading && \
cd ../..Build all solutions:
cmake -B build -D CMAKE_BUILD_TYPE=Release
cmake --build buildRun all tests:
./build/tests/test_common_parts/test_genome_zipper/test_genome_zipper