The official codes for "Towards Building Multilingual Language Model for Medicine".
Models: MMedLM-7B,MMedLM 2-7B,MMedLM 2-1.8B
In this paper, we aim to develop an open-source, multilingual language model for medicine. In general, we present the contribution from the following aspects:
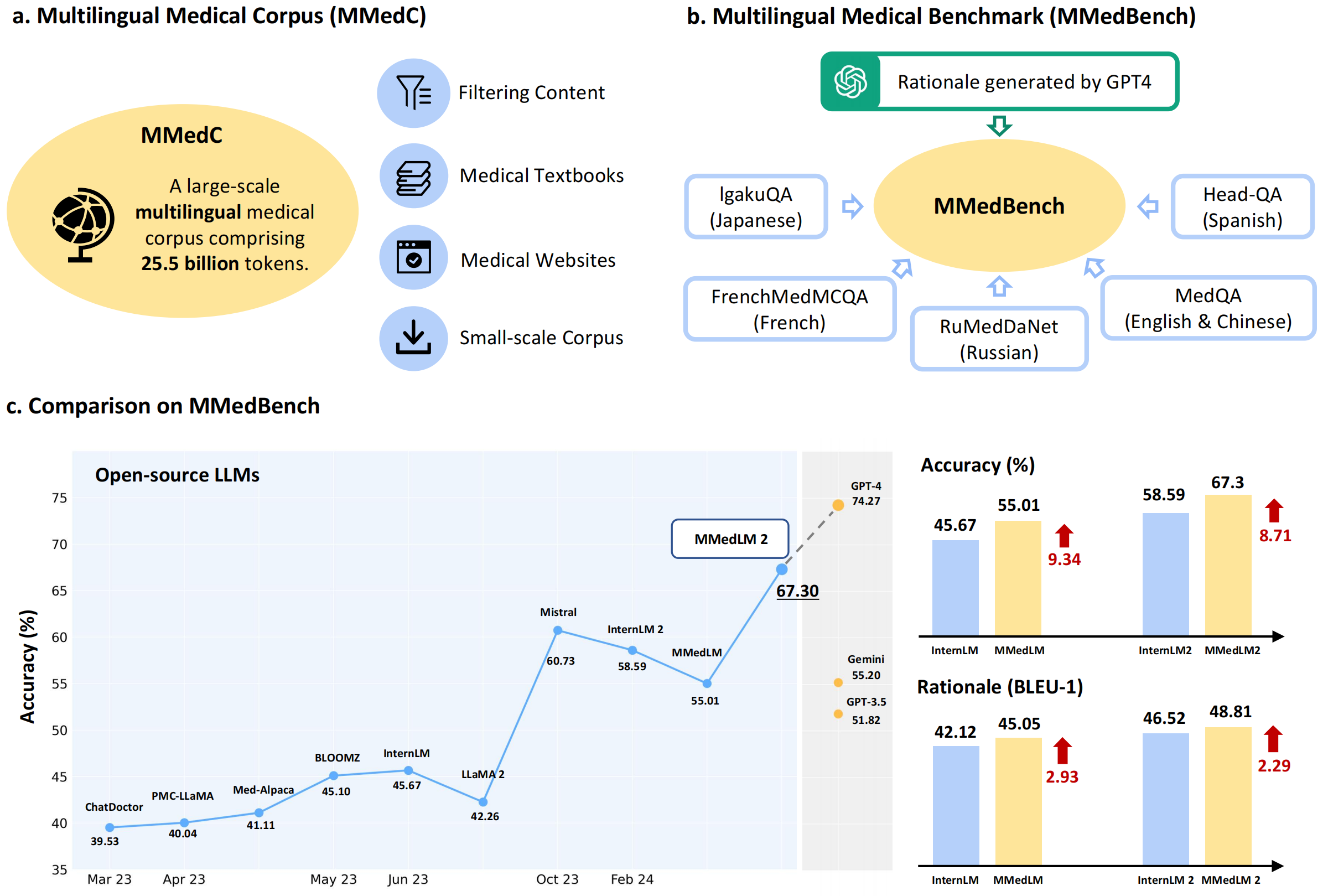
- Corpus dataset. For multilingual medical-specific adaptation, we construct a new multilingual medical corpus, that contains approximately 25.5B tokens encompassing 6 main languages, termed as MMedC, that enables auto-regressive training for existing general LLMs.
- Benchmark. To monitor the development of multilingual LLMs in medicine, we propose a new multilingual medical multi-choice question-answering benchmark with rationale, termed as MMedBench.
- Model Evaluation. We have assessed a number of popular LLMs on our benchmark, along with those further auto-regressive trained on MMedC, as a result, our final model, termed as MMedLM 2, with only 7B parameters, achieves superior performance compared to all other open-source models, even rivaling GPT-4 on MMedBench.
[2023.3.1] We release MMedLM 2-1.8B, a 1.8B light-weight model based on InternLM 2-1.8B. With an auto-regressive continues training on MMedC, MMedLM 2-1.8B can exceed the performance of most 7B models, including InternLM and LLaMA 2.
[2023.2.21] Our leaderboard web can be found here. We look forward to more superior efforts in multilingual medical LLMs!.
[2024.2.21] Our pre-print paper is released ArXiv. Dive into our findings here.
[2024.2.20] We release MMedLM and MMedLM 2. With an auto-regressive continues training on MMedC, these models achieves superior performance compared to all other open-source models, even rivaling GPT-4 on MMedBench.
[2023.2.20] We release MMedC, a multilingual medical corpus containing 25.5B tokens.
[2023.2.20] We release MMedBench, a new multilingual medical multi-choice question-answering benchmark with rationale.
In our experiments, we used A100 80 GB GPUs and the Slurm scheduling system. We provide a Slurm script to launch training. You can also remove the Slurm commands to run the code on a single machine.
For dependencies, we used Pytorch 1.13 and Transformers 4.37. For LoRA fine-tune, it is also necessary to install the corresponding PEFT library.
We provide all the code used for further training on MMedC. The codes are in the pretrain folder. You can check the documentation in the folder for how to use the codes.
- Note that this step requires at least 8 A100 80GB GPUs and training for over a month.
We provide all the code used for fine-tuning. We support 2 fine-tuning methods: Full-Model Fine-tuning and PEFT Fine-Tuning. Both codes are in the finetune folder. You can check the documentation in the folder for how to use the codes.
We provide the code used for inference on MMedBench Testset. The codes are in the pretrain folder. You can check the documentation in the folder for how to use the codes.
We also release our Data Collection Pipeline, including codes of data filtering and Textbooks OCR. For OCR, you may need to install some extra dependencies. Please check out the data_collection folder for more details.
Here we show the main results of models' performance on MMedBench. For more details, please check out our paper.
| Method | Size | Year | MMedC | MMedBench | English | Chinese | Japanese | French | Russian | Spanish | Avg. |
|---|---|---|---|---|---|---|---|---|---|---|---|
| GPT-3.5 | - | 2022.12 | ✗ | ✗ | 56.88 | 52.29 | 34.63 | 32.48 | 66.36 | 66.06 | 51.47 |
| GPT-4 | - | 2023.3 | ✗ | ✗ | 78.00 | 75.07 | 72.91 | 56.59 | 83.62 | 85.67 | 74.27 |
| Gemini-1.0 pro | - | 2024.1 | ✗ | ✗ | 53.73 | 60.19 | 44.22 | 29.90 | 73.44 | 69.69 | 55.20 |
| BLOOMZ | 7B | 2023.5 | ✗ | trainset | 43.28 | 58.06 | 32.66 | 26.37 | 62.89 | 47.34 | 45.10 |
| InternLM | 7B | 2023.7 | ✗ | trainset | 44.07 | 64.62 | 37.19 | 24.92 | 58.20 | 44.97 | 45.67 |
| Llama 2 | 7B | 2023.7 | ✗ | trainset | 43.36 | 50.29 | 25.13 | 20.90 | 66.80 | 47.10 | 42.26 |
| MedAlpaca | 7B | 2023.3 | ✗ | trainset | 46.74 | 44.80 | 29.64 | 21.06 | 59.38 | 45.00 | 41.11 |
| ChatDoctor | 7B | 2023.4 | ✗ | trainset | 43.52 | 43.26 | 25.63 | 18.81 | 62.50 | 43.44 | 39.53 |
| PMC-LLaMA | 7B | 2023.4 | ✗ | trainset | 47.53 | 42.44 | 24.12 | 20.74 | 62.11 | 43.29 | 40.04 |
| Mistral | 7B | 2023.10 | ✗ | trainset | 61.74 | 71.10 | 44.72 | 48.71 | 74.22 | 63.86 | 60.73 |
| InternLM 2 | 1.8B | 2024.2 | ✗ | trainset | 38.49 | 64.1 | 32.16 | 18.01 | 53.91 | 36.83 | 40.58 |
| InternLM 2 | 7B | 2024.2 | ✗ | trainset | 57.27 | 77.55 | 47.74 | 41.00 | 68.36 | 59.59 | 58.59 |
| MMedLM (Ours) | 7B | - | ✓ | trainset | 49.88 | 70.49 | 46.23 | 36.66 | 72.27 | 54.52 | 55.01 |
| MMedLM 2(Ours) | 7B | - | ✓ | trainset | 61.74 | 80.01 | 61.81 | 52.09 | 80.47 | 67.65 | 67.30 |
| MMedLM 2(Ours) | 1.8B | - | ✓ | trainset | 45.40 | 66.78 | 42.21 | 25.56 | 69.14 | 43.40 | 48.75 |
- GPT and Gemini is evluated under zero-shot setting through API
- Open-source models first undergo training on the trainset of MMedBench before evaluate.
| Method | English | Chinese | Japanese | French | Russian | Spanish | Avg. |
|---|---|---|---|---|---|---|---|
| BLOOMZ | 45.94/ 40.51 | 48.37/ 48.26 | 44.71/ 48.61 | 44.47/ 41.05 | 29.95/ 21.50 | 45.91/ 40.77 | 43.22/ 40.12 |
| InternLM | 46.53/ 41.86 | 48.24/ 48.64 | 44.89/ 49.83 | 41.80/ 37.95 | 27.87/ 21.20 | 43.42/ 38.59 | 42.12/ 39.68 |
| Llama 2 | 46.87/ 41.39 | 46.62/ 46.57 | 48.53/ 51.21 | 44.43/ 40.38 | 33.05/ 23.24 | 45.96/ 40.37 | 44.24/ 40.53 |
| MedAlpaca | 47.33/ 42.31 | 45.72/ 46.49 | 45.35/ 49.12 | 43.78/ 40.41 | 32.80/ 23.15 | 45.99/ 40.57 | 43.49/ 40.34 |
| ChatDoctor | 47.22/ 41.97 | 44.66/ 45.81 | 38.87/ 47.95 | 44.64/ 40.25 | 32.19/ 23.37 | 45.68/ 40.71 | 42.21/ 40.01 |
| PMC-LLaMA | 47.33/ 42.87 | 45.87/ 46.18 | 44.52/ 48.44 | 43.80/ 40.23 | 31.14/ 22.28 | 46.30/ 40.68 | 43.16/ 40.12 |
| Mistral | 47.16/ 41.82 | 48.34/ 47.91 | 48.80/ 50.60 | 45.83/ 40.88 | 34.52/ 24.68 | 47.55/ 41.41 | 45.37/ 41.22 |
| InternLM2 | 49.48/ 44.12 | 51.38/ 51.58 | 50.64/ 53.46 | 46.73/ 42.00 | 32.93/ 24.05 | 47.94/ 41.96 | 46.52/ 42.86 |
| MMedLM | 47.37/ 41.98 | 48.68/ 49.28 | 48.95/ 52.34 | 45.39/ 41.41 | 33.24/ 24.67 | 46.68/ 41.35 | 45.05/ 41.84 |
| MMedLM 2 | 50.02/ 44.77 | 51.39/ 51.78 | 54.79/ 57.10 | 49.04/ 45.30 | 37.49/ 28.18 | 50.14/ 44.59 | 48.81/ 45.29 |
- GPT and Gemini is evluated under zero-shot setting through API
- Open-source models first undergo training on the trainset of MMedBench before evaluate.
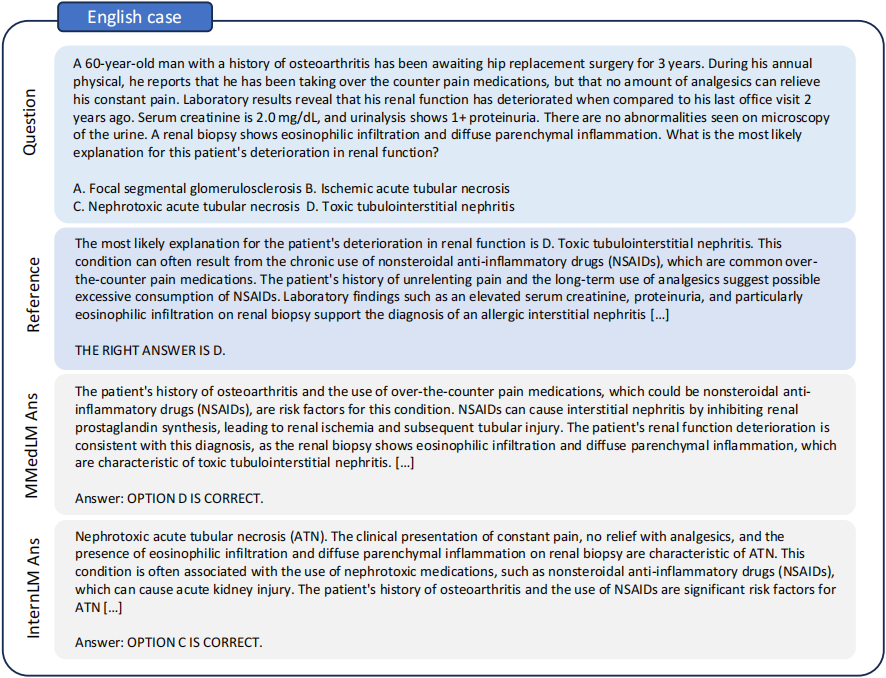
- A case between InternLM and MMedLM. MMedLM demonstrates superior performance in selecting the correct option. Furthermore, MMedLM frequently justifies its choices with accurate reasons. MMedLM accurately diagnoses ‘eosinophilic infiltration in the renal biopsy,’ subsequently applying its domain knowledge to identify these findings as indicative of toxic tubulointerstitial nephritis, leading to a precise diagnosis.
- Add documentation and code for model usage
- Add the Model Card
- Add the Dataset Card
PMC-LLaMA -- https://github.com/chaoyi-wu/PMC-LLaMA
InternLM -- https://github.com/InternLM/InternLM
If you have any question, please feel free to contact qiupengcheng@pjlab.org.cn.
@misc{qiu2024building,
title={Towards Building Multilingual Language Model for Medicine},
author={Pengcheng Qiu and Chaoyi Wu and Xiaoman Zhang and Weixiong Lin and Haicheng Wang and Ya Zhang and Yanfeng Wang and Weidi Xie},
year={2024},
eprint={2402.13963},
archivePrefix={arXiv},
primaryClass={cs.CL}
}