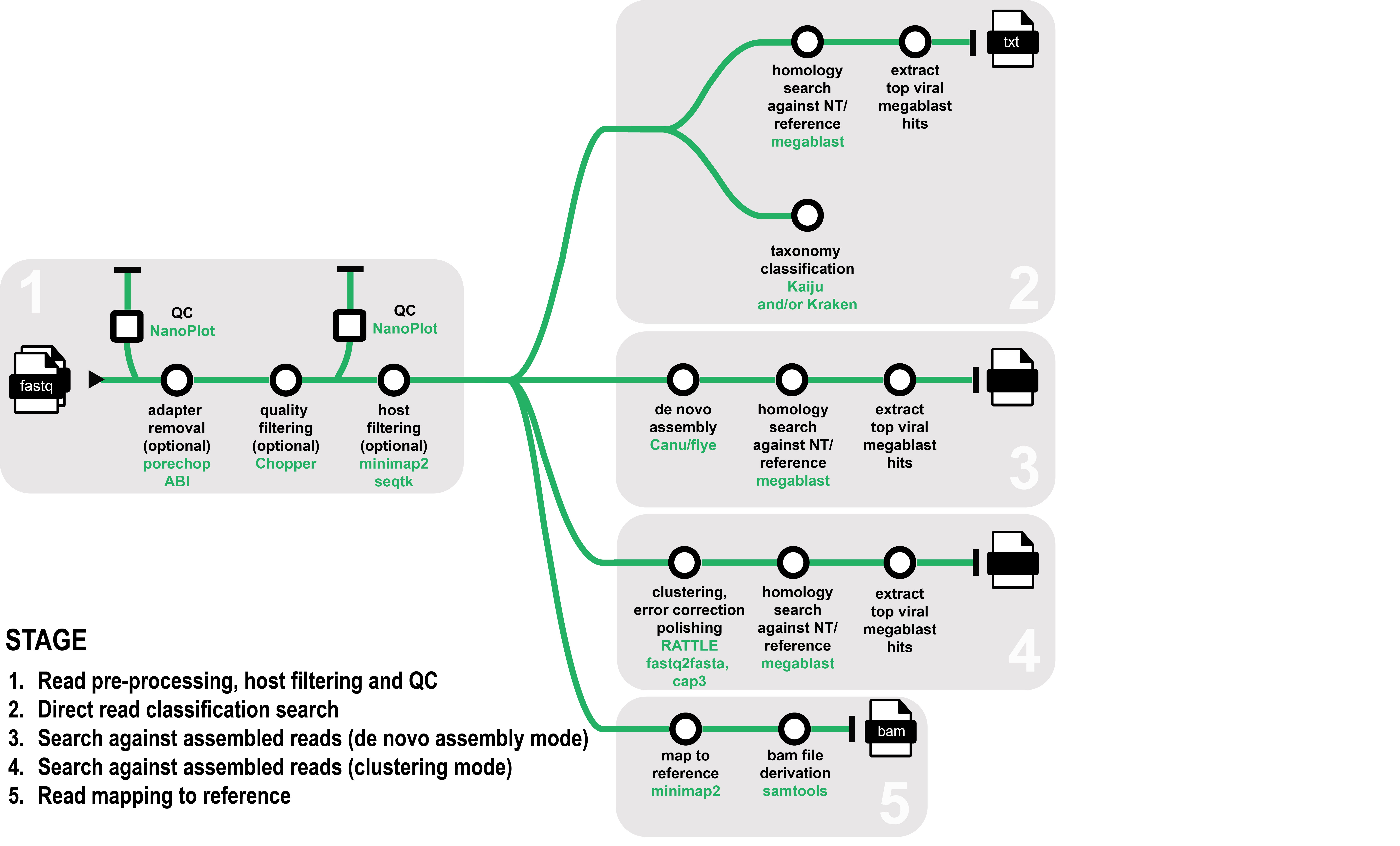
eresearchqut/ontvisc is a Nextflow-based bioinformatics pipeline designed to help diagnostics of viruses and viroid pathogens for biosecurity. It takes fastq files generated from either amplicon or whole-genome sequencing using Oxford Nanopore Technologies as input.
The pipeline can either: 1) perform a direct search on the sequenced reads, 2) generate clusters, 3) assemble the reads to generate longer contigs or 4) directly map reads to a known reference.
The reads can optionally be filtered from a plant host before performing downstream analysis.
- Data quality check (QC) and preprocessing
- Merge fastq files (optional)
- Raw fastq file QC (Nanoplot)
- Trim adaptors (PoreChop ABI - optional)
- Filter reads based on length and/or quality (Chopper - optional)
- Reformat fastq files so read names are trimmed after the first whitespace (bbmap)
- Processed fastq file QC (if PoreChop and/or Chopper is run) (Nanoplot)
- Host read filtering
- Align reads to host reference provided (Minimap2)
- Extract reads that do not align for downstream analysis (seqtk)
- QC report
- Derive read counts recovered pre and post data processing and post host filtering
- Read classification analysis mode
- Clustering mode
- Read clustering (Rattle)
- Convert fastq to fasta format (seqtk)
- Cluster scaffolding (Cap3)
- Megablast homology search against ncbi or custom database (blast)
- Derive top candidate viral hits
- De novo assembly mode
- De novo assembly (Canu or Flye)
- Megablast homology search against ncbi or custom database or reference (blast)
- Derive top candidate viral hits
- Read classification mode
- Option 1 Nucleotide-based taxonomic classification of reads (Kraken2, Braken)
- Option 2 Protein-based taxonomic classification of reads (Kaiju, Krona)
- Option 3 Convert fastq to fasta format (seqtk) and perform direct homology search using megablast (blast)
- Map to reference mode
- Align reads to reference fasta file (Minimap2) and derive bam file and alignment statistics (Samtools)
Detailed instructions can be found in wiki.
- Derive consensus sequence in blast2ref mode
- Finalise output section wiki documentation
- Testing docker mode
- issue with single Chopper params definition requiring a space
Marie-Emilie Gauthier gauthiem@qut.edu.au
Craig Windell c.windell@qut.edu.au
Magdalena Antczak magdalena.antczak@qcif.edu.au
Roberto Barrero roberto.barrero@qut.edu.au