🐧 WIP
Install Anaconda for your operating system (Choosing the Graphical or Command Line Installer depends on your preference).
Brew is a Package Manager for MacOS that can be installed with these instructions. To install, open Terminal and copy+paste:
$ /bin/bash -c "$(curl -fsSL https://raw.githubusercontent.com/Homebrew/install/HEAD/install.sh)"Once Brew is installed, you can install OpenSlide via:
$ brew install openslide$ conda create -n patch-viewer python=3.9
$ conda activate patch-viewer
$ pip install "napari[all]" # install napari
$ pip install git+git://github.com/manzt/patch-viewer.git # install this package from githubTo Open Napari:
$ napari # opens viewer, can drag and drop files
# or
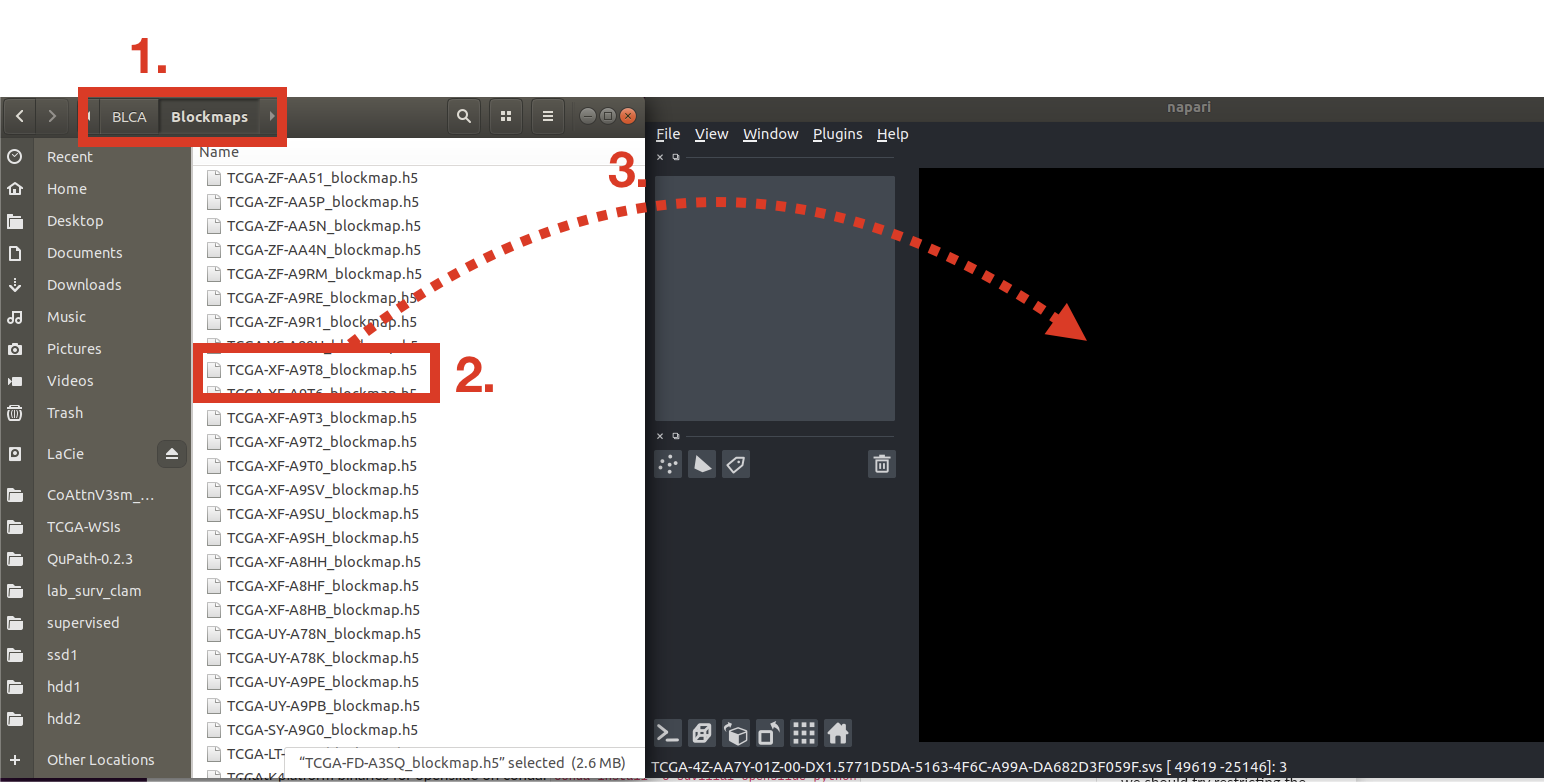
$ napari path/to/my_blockmap.h5 # automatically loads view for fileEach dataset folder contains: 1) a Raw_HE folder containing the raw H&E images, 2) Blockmaps folder containing the heatmaps. To open a heatmap,
- Go to the Blockmaps directory
- Find the .h5 file you are interested in visualizing
- Drag-and-drop the .h5 file to Napari
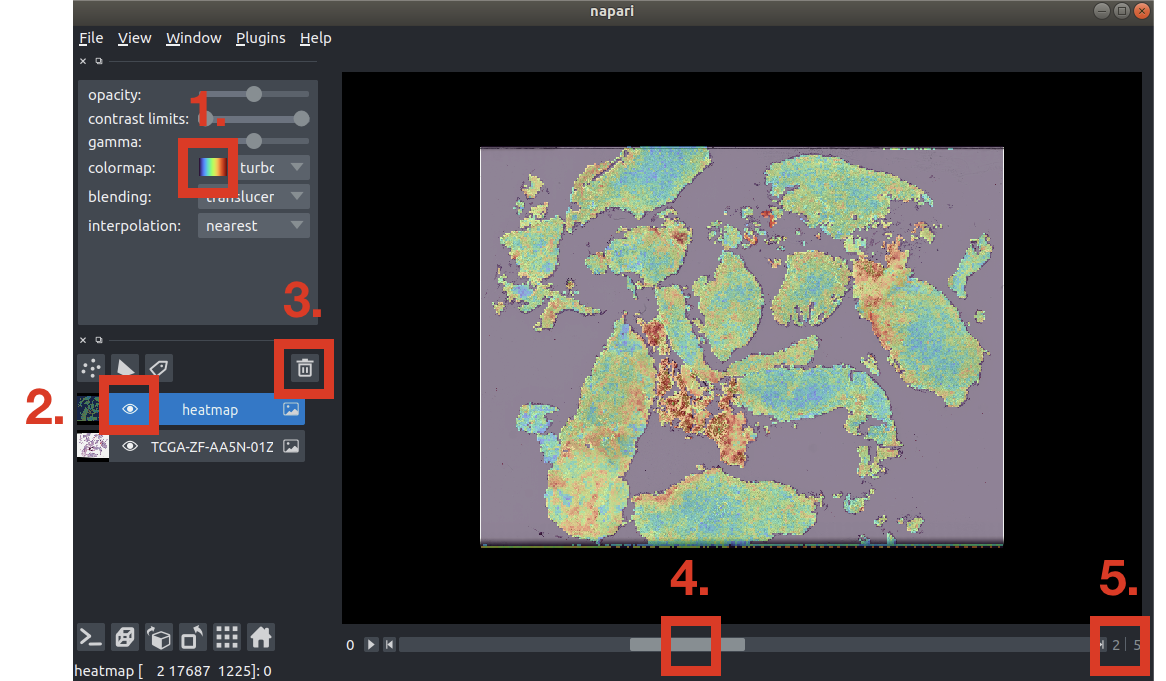
Below is a walkthrough on the UI of Napari:
- Selects the desired colormap. "turbo" is set as the default.
- Similar to Adobe Photoshop, images are opened as masks. To switch between H&E and the heatmaps, click on the "hide" button to deselect vieweing the heatmap mask.
- Before opening another .h5 file, delete the currently opened masks.
- Slider bar to switch between the co-attention heatmaps.
- Counter on the current heatmap you are viewing.
Co-Attention Heatmap Signature Names:
- Tumor Supressor Genes
- Oncogenes
- Protein Kinases
- Cell Differentiation Markers
- Transcription Factors
- Cyokines and Growth Factors