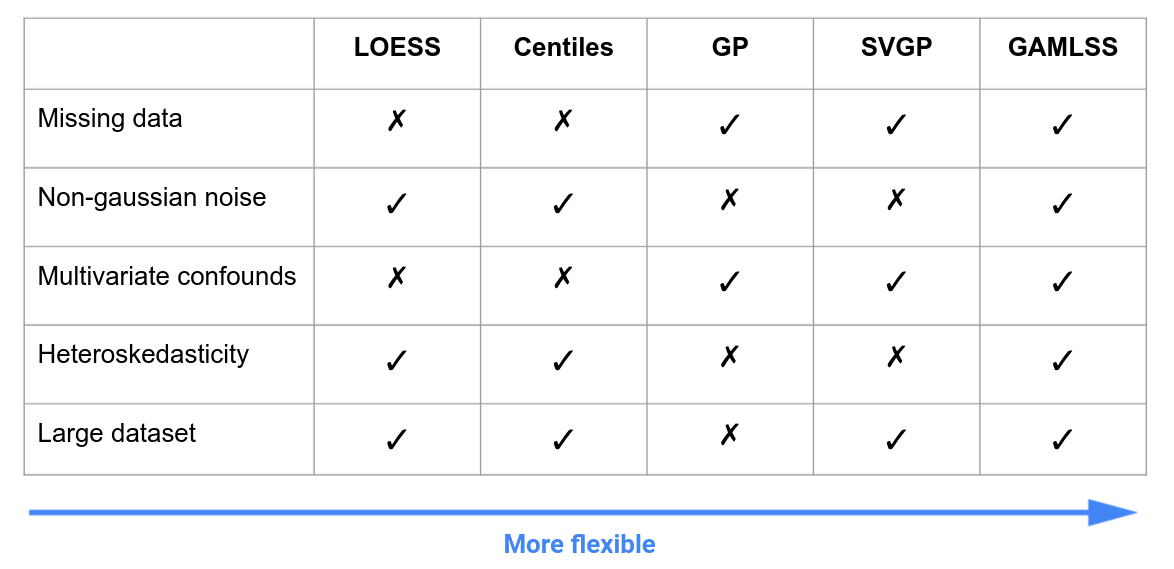
PyNM is a lightweight python implementation of Normative Modeling making it approachable and easy to adopt. The package provides:
- Python API and a command-line interface for wide accessibility
- Automatic dataset splitting and cross-validation
- Five models from various back-ends in a unified interface that cover a broad range of common use cases
- Centiles
- LOESS
- Gaussian Process (GP)
- Stochastic Variational Gaussian Process (SVGP)
- Generalized Additive Models of Location Shape and Scale (GAMLSS)
- Solutions for very large datasets and heteroskedastic data
- Integrated plotting and evaluation functions to quickly check the validity of the model fit and results
- Comprehensive and interactive tutorials
The tutorials can be accessed without any local installation via binder:
For a more advanced implementation, see the Python library PCNtoolkit.
Note: functional installation requires python 3.9
Minimal Installation (without R)
If you aren't using the GAMLSS model/don't need to install R.
$ pip install pynmInstallation with R
If you are using a GAMLSS.
- Must first have R (v4.2.2) installed and packages:
- gamlss
- gamlss.dist
- gamlss.data
Instruction for installing R can be found at r-project. Once R and the gamlss packages are installed, install pynm:
$ pip install pynmBleeding-edge Installation
If you want to be up to date with the most recent changes to PyNM (not necessarily stable). For the options above replace pip install pynm with:
$ git clone https://github.com/ppsp-team/PyNM.git
$ cd pynm
$ pip install .usage: pynm [-h] --pheno_p PHENO_P --out_p OUT_P --confounds CONFOUNDS --score
SCORE --group GROUP [--train_sample TRAIN_SAMPLE] [--LOESS]
[--centiles] [--bin_spacing BIN_SPACING] [--bin_width BIN_WIDTH]
[--GP] [--gp_method GP_METHOD] [--gp_num_epochs GP_NUM_EPOCHS]
[--gp_n_inducing GP_N_INDUCING] [--gp_batch_size GP_BATCH_SIZE]
[--gp_length_scale GP_LENGTH_SCALE]
[--gp_length_scale_bounds [GP_LENGTH_SCALE_BOUNDS [GP_LENGTH_SCALE_BOUNDS ...]]]
[--gp_nu NU] [--GAMLSS] [--gamlss_mu GAMLSS_MU]
[--gamlss_sigma GAMLSS_SIGMA] [--gamlss_nu GAMLSS_NU]
[--gamlss_tau GAMLSS_TAU] [--gamlss_family GAMLSS_FAMILY]
optional arguments:
-h, --help show this help message and exit
--pheno_p PHENO_P Path to phenotype data. Data must be in a .csv file.
--out_p OUT_P Path to output directory.
--confounds CONFOUNDS
List of confounds to use in the GP model.The list must
formatted as a string with commas between confounds,
each confound must be a column name from the phenotype
.csv file. For GP model all confounds will be used,
for LOESS and Centiles models only the first is used.
For GAMLSS all confounds are used unless formulas are
specified. Categorical values must be denoted by
c(var) ('c' must be lower case), e.g. 'c(SEX)' for
column name 'SEX'.
--score SCORE Response variable for all models. Must be a column
title from phenotype .csv file.
--group GROUP Column name from the phenotype .csv file that
distinguishes probands from controls. The column must
be encoded with str labels using 'PROB' for probands
and 'CTR' for controls or with int labels using 1 for
probands and 0 for controls.
--train_sample TRAIN_SAMPLE
Which method to use for a training sample, can be a
float in (0,1] for a percentage of controls or
'manual' to be manually set using a column of the
DataFrame labelled 'train_sample'.
--LOESS Flag to run LOESS model.
--centiles Flag to run Centiles model.
--bin_spacing BIN_SPACING
Distance between bins for LOESS & centiles models.
--bin_width BIN_WIDTH
Width of bins for LOESS & centiles models.
--GP Flag to run Gaussian Process model.
--gp_method GP_METHOD
Method to use for the GP model. Can be set to
'auto','approx' or 'exact'. In 'auto' mode, the exact
model will be used for datasets smaller than 2000 data
points. SVGP is used for the approximate model. See
documentation for details. Default value is 'auto'.
--gp_num_epochs GP_NUM_EPOCHS
Number of training epochs for SVGP model. See
documentation for details. Default value is 20.
--gp_n_inducing GP_N_INDUCING
Number of inducing points for SVGP model. See
documentation for details. Default value is 500.
--gp_batch_size GP_BATCH_SIZE
Batch size for training and predicting from SVGP
model. See documentation for details. Default value is
256.
--gp_length_scale GP_LENGTH_SCALE
Length scale of Matern kernel for exact model. See
documentation for details. Default value is 1.
--gp_length_scale_bounds [GP_LENGTH_SCALE_BOUNDS [GP_LENGTH_SCALE_BOUNDS ...]]
The lower and upper bound on length_scale. If set to
'fixed', length_scale cannot be changed during
hyperparameter tuning. See documentation for details.
Default value is (1e-5,1e5).
--gp_nu NU Nu of Matern kernel for exact and SVGP model. See
documentation for details. Default value is 2.5.
--GAMLSS Flag to run GAMLSS.
--gamlss_mu GAMLSS_MU
Formula for mu (location) parameter of GAMLSS. Default
formula for score is sum of confounds with non-
categorical columns as smooth functions, e.g. 'score ~
ps(age) + sex'.
--gamlss_sigma GAMLSS_SIGMA
Formula for mu (location) parameter of GAMLSS. Default
formula is '~ 1'.
--gamlss_nu GAMLSS_NU
Formula for mu (location) parameter of GAMLSS. Default
formula is '~ 1'.
--gamlss_tau GAMLSS_TAU
Formula for mu (location) parameter of GAMLSS. Default
formula is '~ 1'.
--gamlss_family GAMLSS_FAMILY
Family of distributions to use for fitting, default is
'SHASHo2'. See R documentation for GAMLSS package for
other available families of distributions.
from pynm.pynm import PyNM
# Load data
df = pd.read_csv('data.csv')
# Initialize pynm w/ data and confounds
m = PyNM(df,'score','group', confounds = ['age','c(sex)','c(site)'])
# Run models
m.loess_normative_model()
m.centiles_normative_model()
m.gp_normative_model()
m.gamlss_normative_model()
# Collect output
data = m.dataAll the functions have the classical Python DocStrings that you can summon with help(). You can also see the tutorials for documented examples.
By default, the models are fit on all the controls in the dataset and prediction is then done on the entire dataset. The residuals (scores of the normative model) are then calculated as the difference between the actual value and predicted value for each subject. This paradigm is not meant for situations in which the residuals will then be used in a prediction setting, since any train/test split stratified by proband/control will have information from the training set leaked into the test data.
In order to avoid contaminating the test set, in a prediction setting it is important to fit the normative model on a subset of the controls and then leave those out. This is implemented in PyNM with the --train_sample flag. It can be set to:
- A number in (0,1]
- This is simplest usage that defines the sample size, PyNM will then select a random sample of the controls and use those as a training group. The number is the proportion of controls to use, the default value is 1 to use the full set of controls.
- The subjects used in the sample are recorded in the column
'train_sample'of the resulting PyNM.data object. Subjects used in the training sample are encoded as 1s, and the rest as 0s.
'manual'- It is also possible to specify exactly which subjects to use as a training group by providing a column in the input data labeled
'train_sample'encoded the same way.
- It is also possible to specify exactly which subjects to use as a training group by providing a column in the input data labeled
Both the Centiles and LOESS models are non parametric models based local approximations. They accept only a single dependent variable, passed using the conf option.
Gaussian Process Regression (GPR), which underpins the Gaussian Process Model, can accept an arbitrary number of dependent variables passed using the confounds option. Note: in order for GPR to be effective, the data must be homoskedastic. For a full discussion see this paper.
GPR is very intensive on both memory and time usage. In order to have a scaleable method, we've implemented both an exact model for smaller datasets and an approximate method, recommended for datasets over ~1000 subjects. The method can be specified using the method option, it defaults to auto in which the approxiamte model will be chosen for datasets over 1000.
The exact model implements scikit-learn's Gaussian Process Regressor. The kernel is composed of a constant kernel, a white noise kernel, and a Matern kernel. The Matern kernel has parameters nu and length_scale that can be specified. The parameter nu has special values at 1.5 and 2.5, using other values will significantly increase computation time. See documentation for an overview of both.
The approximate model implements a Stochastic Variational Gaussian Process (SVGP) model using GPytorch, with a kernel closely matching the one in the exact model. SVGP is a deep learning technique that needs to be trained on minibatches for a set number of epochs, this can be tuned with the parameters batch_size and num_epoch. The model speeds up computation by using a subset of the data as inducing points, this can be controlled with the parameter n_inducing that defines how many points to use. See documentation for an overview.
Generalized Additive Models of Location Shape and Scale (GAMLSS) are a flexible modeling framework that can model heteroskedasticity, non-linear effects of variables, and hierarchical structure of the data. The implementation here is a python wrapper for the R package gamlss, formulas for each parameter must be specified using functions available in the package (see documentation). For a full discussion of using GAMLSS for normative modeling see this paper.
Original papers with Gaussian Processes (GP):
- Marquand et al. Biological Psychiatry 2016 doi:10.1016/j.biopsych.2015.12.023
- Marquand et al. Molecular Psychiatry 2019 doi:10.1038/s41380-019-0441-1
For limitations of Gaussian Proccesses:
- Xu et al. PLoS ONE 2021, The pitfalls of using Gaussian Process Regression for normative modeling
Example of use of the LOESS approach:
- Lefebvre et al. Front. Neurosci. 2018 doi:10.3389/fnins.2018.00662
- Maruani et al. Front. Psychiatry 2019 doi:10.3389/fpsyt.2019.00011
For the Centiles approach see:
- Bethlehem et al. Communications Biology 2020 doi:10.1038/s42003-020-01212-9
- R implementation here.
For the SVGP model see:
- Hensman et al. https://arxiv.org/pdf/1411.2005.pdf
For GAMLSS see:
- Dinga et al. https://doi.org/10.1101/2021.06.14.448106
- R documentation https://cran.r-project.org/web/packages/gamlss/index.html
To test the code locally, first make sure R and the required packages are installed then follow the instructions above under Installation: Bleeding-edge Installation. Finally, run:
$ pip install -r requirements.txt
$ pytest test/test_pynm.pyIf you spot any bugs 🪲? Check out the open issues to see if we're already working on it. If not, open up a new issue and we will check it out when we can!
Thank you for considering contributing to our project! Before getting involved, please review our contribution guidelines.
This work is supported by IVADO, FRQS, CFI, MITACS, and Compute Canada.