This repository provides the code for "Uncertainty-Guided Efficient Interactive Refinement of Fetal Brain Segmentation from Stacks of MRI Slices" accepted by MICCAI 2020. The paper can be found at: https://arxiv.org/abs/2007.00833.
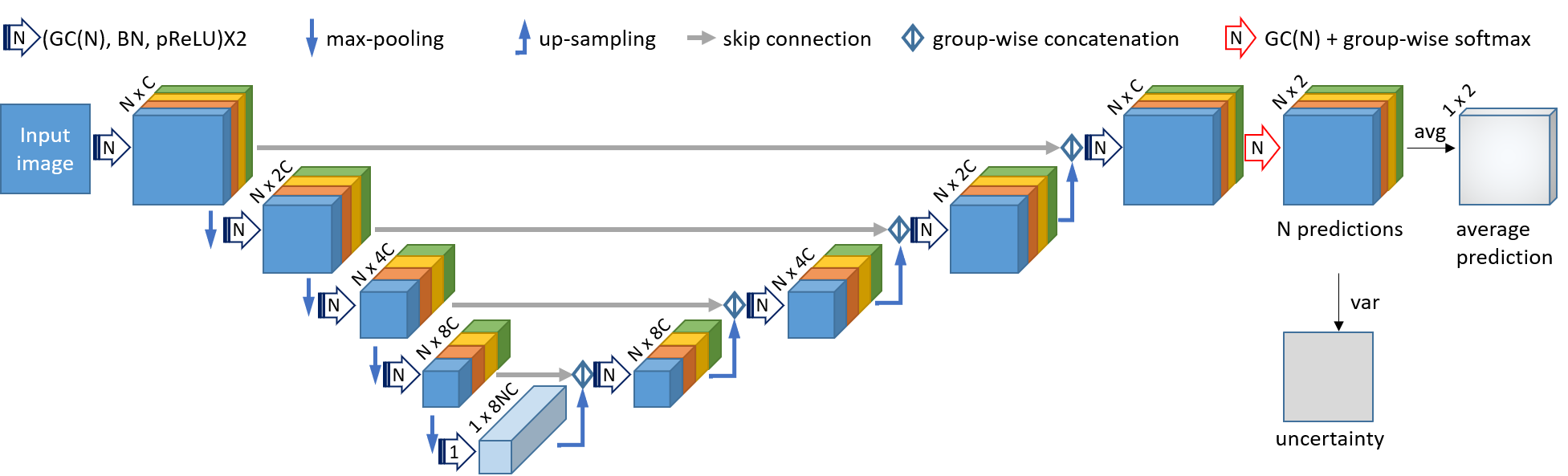
The code contains two modules: 1), a novel CNN based on convolution in Multiple Groups (MG-Net) that simultaneously obtains an intial segmentation and its uncertainty estimation. 2), Interaction-based level set for fast refinement, which is an extention of the DRLSE algorithm and named as I-DRLSE.
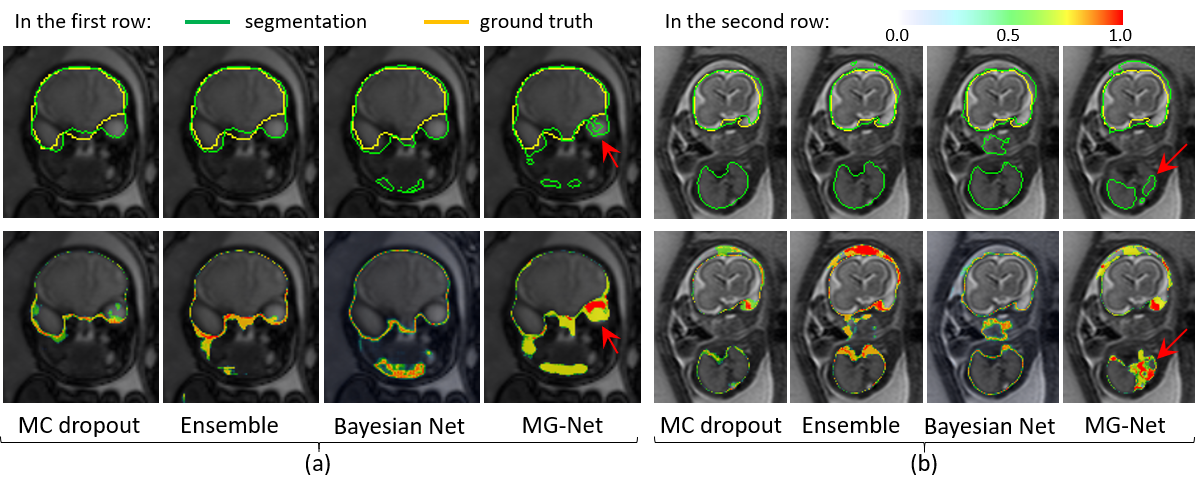 Fig. 2. Segmentation with uncertainty estimation.
Fig. 2. Segmentation with uncertainty estimation.
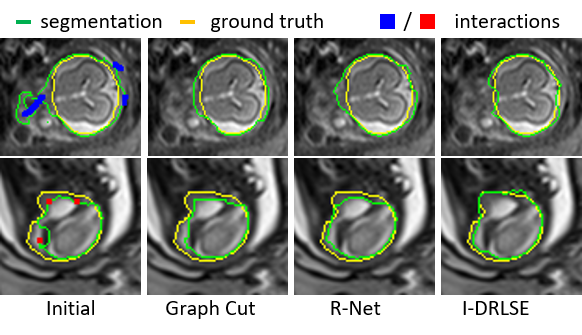
Fig. 3. Using I-DRLSE for interactive refinement.
Some important required packages include:
- Pytorch version >=1.0.1.
- PyMIC, a pytorch-based toolkit for medical image computing. Version 0.2 is required.
- GeodisTK, geodesic distance transform toolkit for 2D and 3D images.
Follow official guidance to install Pytorch. Install the other required packages by:
pip install -r requirements.txt
After installing the required packages, add the path of UGIR to the PYTHONPATH environment variable.
- Run the following commands to use MG-Net for simultanuous segmentation and uncertainty estimation.
cd uncertainty_demo
python ../util/custom_net_run.py test config/mgnet.cfg
- The results will be saved to
uncertainty_demo/result. To get a visualization of the uncertainty estimation in an example slice, run:
python show_uncertanty.py
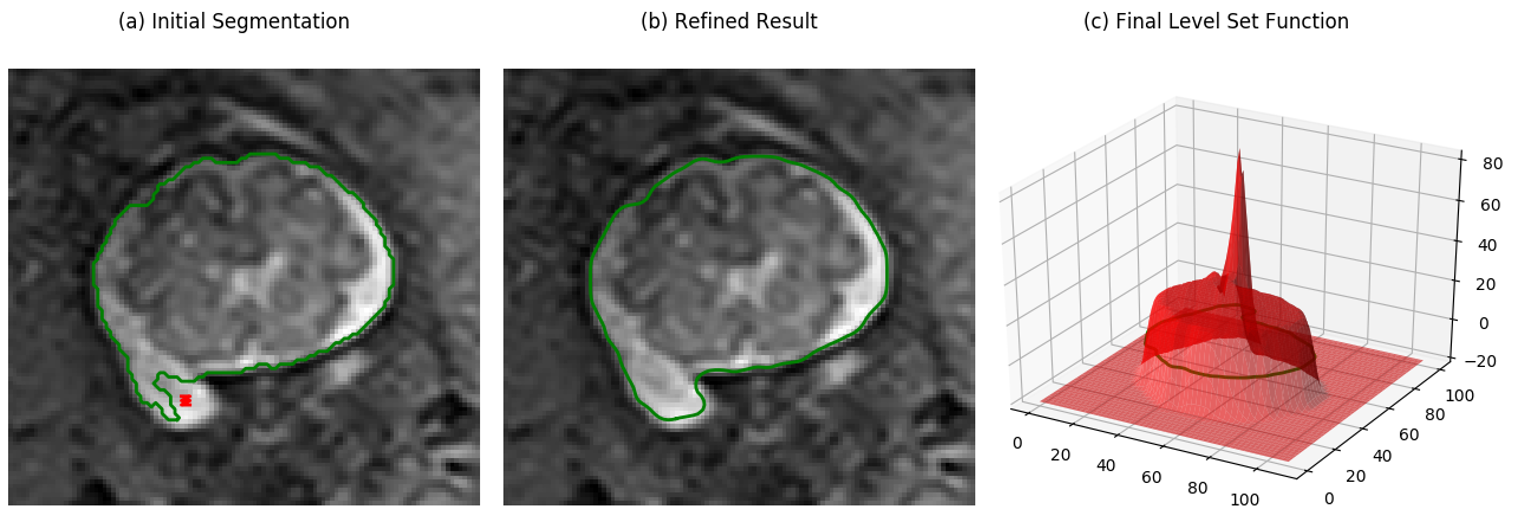
To see a demo of I-DRLSE, run the following commands:
cd util/level_set
python demo/demo_idrlse.py
The result should look like the following.
Copyright (c) 2020, University of Electronic Science and Technology of China. All rights reserved. This code is made available as open-source software under the BSD-3-Clause License.