NeuMiss
This repo provides a PyTorch module implementing NeuMiss, a neural network architecture designed for learning with missing values.
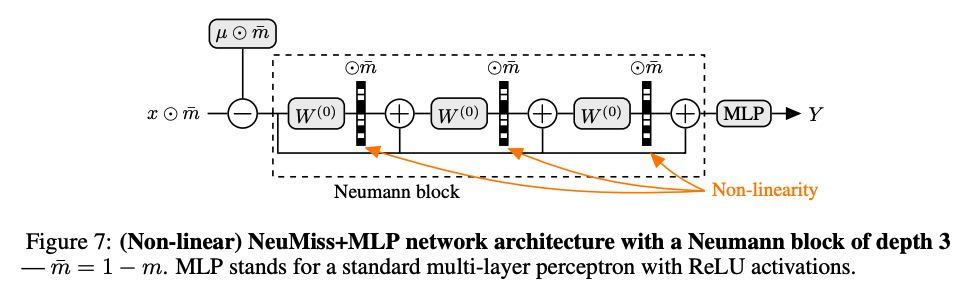
The NeuMiss block is implemented in NeuMissBlock. It takes as input a vector with missing values and outputs a vector of the same size as the input, without mising values. It is thus necessary to chain it with subsequent layers appropriate to the prediction task. As an example we provide NeuMissMLP, where the NeuMiss block is chained with an MLP. The NeuMissBlock can be used like any other PyTorch layer, and can easily be chained with other architectures (ResNets, ...).
Below is a diagram of a NeuMiss block chained with an MLP:
For more details, please refer to the papers introducing NeuMiss:
The architecture implemented corresponds to the latest version (i.e. with shared weights) introduced in the second paper.
Install
Clone this repository and install it with pip in the environment of your choice (venv, conda...):
pip install .Use NeuMiss
Below, we show how to train NeuMissMLP on a simple synthetic dataset.
Go to src folder and import the required modules.
cd src
import torch.nn as nn
from torch.utils.data import DataLoader
from torch.optim.lr_scheduler import ReduceLROnPlateau
from neumiss import NeuMissMLP
from generate_example_dataset import get_example_dataset
from utils import get_optimizer_by_group, train_model, \
compute_preds, compute_regression_metricsGenerate a synthetic dataset. Here, we generate Gaussian data (10,000 samples, 10 features) with 50% MCAR missing values. The response is generated linearly from the complete data. Note that with NeuMiss, it is not necessary to impute the data. The NeuMissBlock expects and handles NaN in the inputs.
ds_train, ds_val, ds_test = get_example_dataset()
p = ds_train.tensors[0].shape[1] # n_features
train_loader = DataLoader(ds_train, batch_size=200, shuffle=True)
val_loader = DataLoader(ds_val, batch_size=200)
test_loader = DataLoader(ds_test, batch_size=200)Instantiate a NeuMissMLP network.
model = NeuMissMLP(n_features=p, neumiss_depth=10, mlp_depth=0, mlp_width=p)Instantiate an optimizer, a scheduler and a loss.
optim_hyperparams = {'weight_decay': 0, 'lr': 1e-3}
optimizer = get_optimizer_by_group(model, optim_hyperparams)
sched_hyperparams = {'factor': 0.2, 'patience': 10, 'threshold': 1e-4}
scheduler = ReduceLROnPlateau(optimizer, mode='min', **sched_hyperparams)
criterion = nn.MSELoss()Train the model.
train_model(model, criterion, train_loader, val_loader, optimizer,
scheduler, early_stopping=False, n_epochs=500, lr_threshold=1e-6)Compute prediction scores.
train_loader = DataLoader(ds_train, batch_size=200, shuffle=False)
pred = compute_preds(model, train_loader, val_loader, test_loader,
classif=False)
res = {}
splits = ['train', 'val', 'test']
preds = [pred[split] for split in splits]
y_labels = [ds_train.tensors[1], ds_val.tensors[1], ds_test.tensors[1]]
for split, pred, y_label in zip(splits, preds, y_labels):
res_split = compute_regression_metrics(pred, y_label)
for metric, value in res_split.items():
res[f'{metric}_{split}'] = value
print(res)