# Clone the repository
git clone https://github.com/mariusrueve/ChemWeaver.git
# Change directory
cd ChemWeaver
# Install dependencies
pip install -r requirements.txt
# Run docker-compose
docker-compose up -dNow wait for the database to start up. You can check if the database is ready by accessing the application at http://localhost:7474. The default username and password are neo4j and 1234.
# Run the application
python main.pyAfter running the Python script, the molecules will be loaded into the database and the similarities will be calculated. The results can be accessed by querying the database with Cypher.
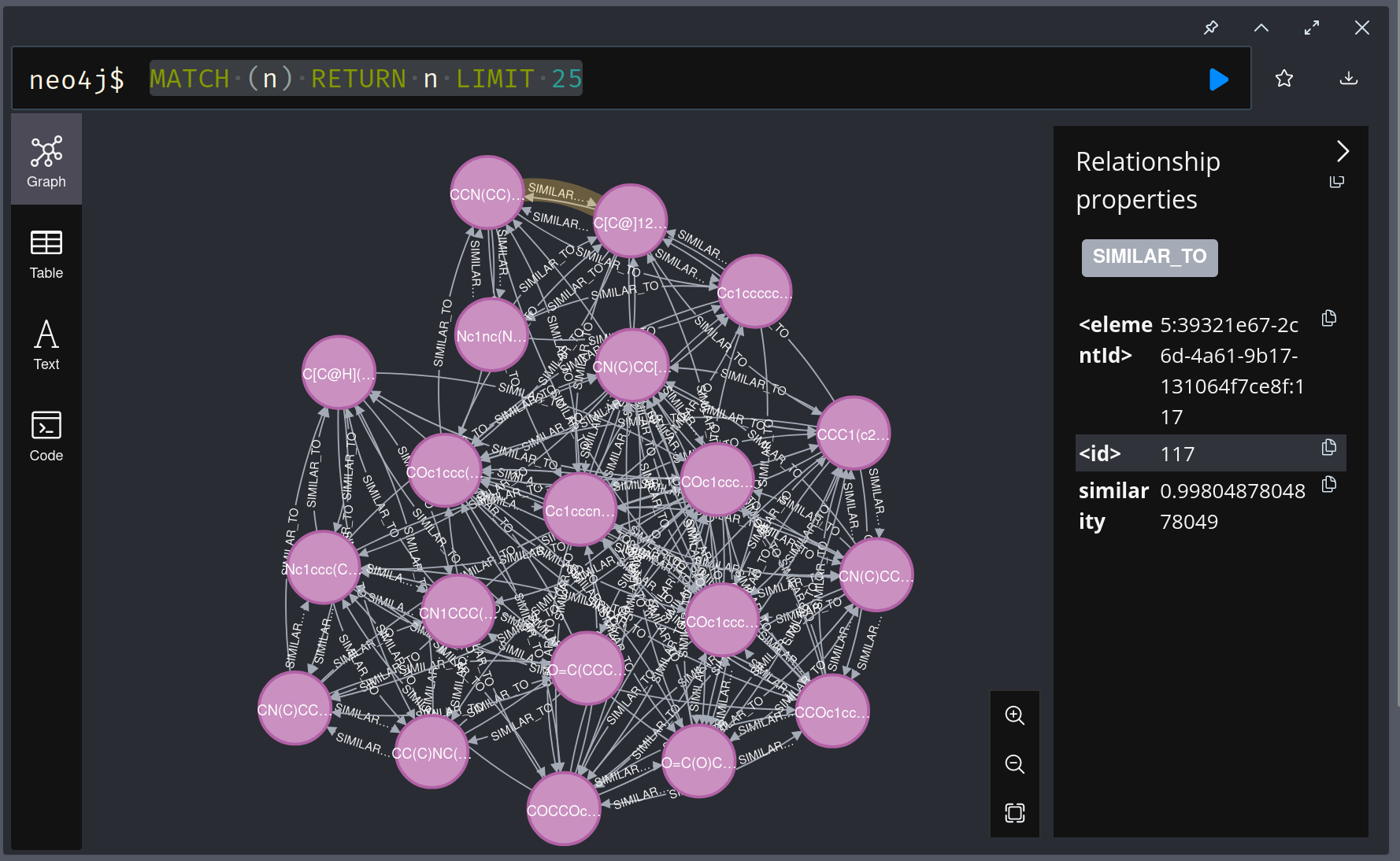
MATCH (n) RETURN n LIMIT 25The results will look like this:
Creating a chemical similarity database using a graph-based database along with fingerprints and other descriptors is a great way to manage and analyze chemical data efficiently. Here’s a step-by-step guide to get you started:
First, select a suitable graph database. Neo4j and ArangoDB are popular choices due to their strong querying capabilities and support for complex data structures.
Chemical structures can be represented as graphs where nodes represent atoms, and edges represent bonds. You will also need to decide on the chemical descriptors:
- Fingerprints: Common types include MACCS keys, ECFP (Extended Connectivity Fingerprints), and RDKit fingerprints. These are used to quickly estimate similarity between molecules.
- Other Descriptors: These might include molecular weight, logP, or specific structural features. Tools like RDKit or Open Babel can calculate these descriptors.
- Extract chemical data: Source your chemical data from databases like PubChem or ChemSpider, or use datasets provided by your institution or company.
- Process data with a cheminformatics tool: Use tools like RDKit in Python to generate fingerprints and calculate other descriptors. For each molecule, you will:
- Compute the fingerprint and convert it to a suitable format for the graph database.
- Calculate other desired descriptors.
- Create nodes and edges: Each molecule can be a node with edges representing bonds or relationships (like similarity scores) to other molecules.
- Store descriptors as properties: Node properties can include fingerprints and other descriptors. You might store fingerprints as bit strings or hash codes.
- Similarity Queries: Implement functions to compare fingerprints using similarity coefficients (Tanimoto, Dice, etc.). This can often be done within the database using custom scripts or external calls to a cheminformatics toolkit.
- Graph Queries: Use Cypher (for Neo4j) or AQL (for ArangoDB) to query molecules based on structural features or calculated properties.
Create indexes on frequently searched properties like molecular weight or specific substructures to speed up query performance.
Develop an API to interact with your database, allowing users to submit queries and retrieve results programmatically. This can be built using frameworks like Flask for Python.
Consider building a user interface that allows users to visualize query results, perhaps showing molecular structures or graphs of related compounds. Tools like ChemDoodle or JSmol can be integrated into web applications.
Implement security measures to control access to the database, ensuring that sensitive data is protected and access is logged.
Regularly test the database with known chemical queries to ensure accuracy and performance. Validate the similarity measures by comparing them with established benchmarks or literature values.
By following these steps, you can build a robust chemical similarity database using a graph-based approach, leveraging the power of fingerprints and other molecular descriptors to provide insightful and fast chemical data analysis.