- Introduction
- Example: A rule ensemble for predicting ozone levels
- Tools for interpretation
- Tuning parameters of function pre
- Dealing with missing values
- Go sparser with relaxed lasso fits
- Generalized Prediction Ensembles: Combining MARS, rules and linear terms
- Credits
- References
pre is an R package for deriving prediction rule ensembles for
binary, multinomial, (multivariate) continuous, count and survival
responses. Input variables may be numeric, ordinal and categorical. An
extensive description of the implementation and functionality is
provided in Fokkema (2020). The package largely implements the algorithm
for deriving prediction rule ensembles as described in Friedman &
Popescu (2008), with several adjustments:
- The package is completely
Rbased, allowing users better access to the results and more control over the parameters used for generating the prediction rule ensemble. - The unbiased tree induction algorithms of Hothorn, Hornik, & Zeileis (2006) is used for deriving prediction rules, by default. Alternatively, the (g)lmtree algorithm of Zeileis, Hothorn, & Hornik (2008) can be employed, or the classification and regression tree (CART) algorithm of Breiman, Friedman, Olshen, & Stone (1984).
- The package supports a wider range of response variable types.
- The initial ensemble may be generated as a bagged, boosted and/or random forest ensemble.
- Hinge functions of predictor variables may be included as
baselearners, as in the multivariate adaptive regression splines
(MARS) approach of Friedman (1991), using function
gpe(). - Tools for explaining individual predictions are provided.
Note that pre is under development, and much work still needs to
be done. Below, an introduction the the package is provided. Fokkema
(2020) provides an extensive description of the fitting procedures
implemented in function pre() and example analyses with more extensive
explanations. An extensive introduction aimed at researchers in social
sciences is provided in Fokkema & Strobl (2020).
To get a first impression of how function pre() works, we will predict
Ozone levels using the airquality dataset. We fit a prediction rule
ensemble using function pre():
library("pre")
airq <- airquality[complete.cases(airquality), ]
set.seed(42)
airq.ens <- pre(Ozone ~ ., data = airq)Note that it is necessary to set the random seed, to allow for later replication of results, because the fitting procedure depends on random sampling of training observations.
We can print the resulting ensemble (alternatively, we could use the
print method):
airq.ens
#>
#> Final ensemble with cv error within 1se of minimum:
#>
#> lambda = 3.543968
#> number of terms = 12
#> mean cv error (se) = 352.395 (99.13754)
#>
#> cv error type : Mean-Squared Error
#>
#> rule coefficient description
#> (Intercept) 68.48270407 1
#> rule191 -10.97368180 Wind > 5.7 & Temp <= 87
#> rule173 -10.90385519 Wind > 5.7 & Temp <= 82
#> rule42 -8.79715538 Wind > 6.3 & Temp <= 84
#> rule204 7.16114781 Wind <= 10.3 & Solar.R > 148
#> rule10 -4.68646145 Temp <= 84 & Temp <= 77
#> rule192 -3.34460038 Wind > 5.7 & Temp <= 87 & Day <= 23
#> rule51 -2.27864287 Wind > 5.7 & Temp <= 84
#> rule93 2.18465676 Temp > 77 & Wind <= 8.6
#> rule74 -1.36479545 Wind > 6.9 & Temp <= 84
#> rule28 -1.15326093 Temp <= 84 & Wind > 7.4
#> rule25 -0.70818400 Wind > 6.3 & Temp <= 82
#> rule166 -0.04751152 Wind > 6.9 & Temp <= 82The first few lines of the printed results provide the penalty parameter
value (λ) employed for selecting the final ensemble. By default, the
‘1-SE’ rule is used for selecting λ; this default can be overridden by
employing the penalty.par.val argument of the print method and other
functions in the package. Note that the printed cross-validated error is
calculated using the same data as was used for generating the rules and
likely provides an overly optimistic estimate of future prediction
error. To obtain a more realistic prediction error estimate, we will use
function cvpre() later on.
Next, the printed results provide the rules and linear terms selected in
the final ensemble, with their estimated coefficients. For rules, the
description column provides the conditions. For linear terms (which
were not selected in the current ensemble), the winsorizing points used
to reduce the influence of outliers on the estimated coefficient would
be printed in the description column. The coefficient column
presents the estimated coefficient. These are regression coefficients,
reflecting the expected increase in the response for a unit increase in
the predictor, keeping all other predictors constant. For rules, the
coefficient thus reflects the difference in the expected value of the
response when the conditions of the rule are met, compared to when they
are not.
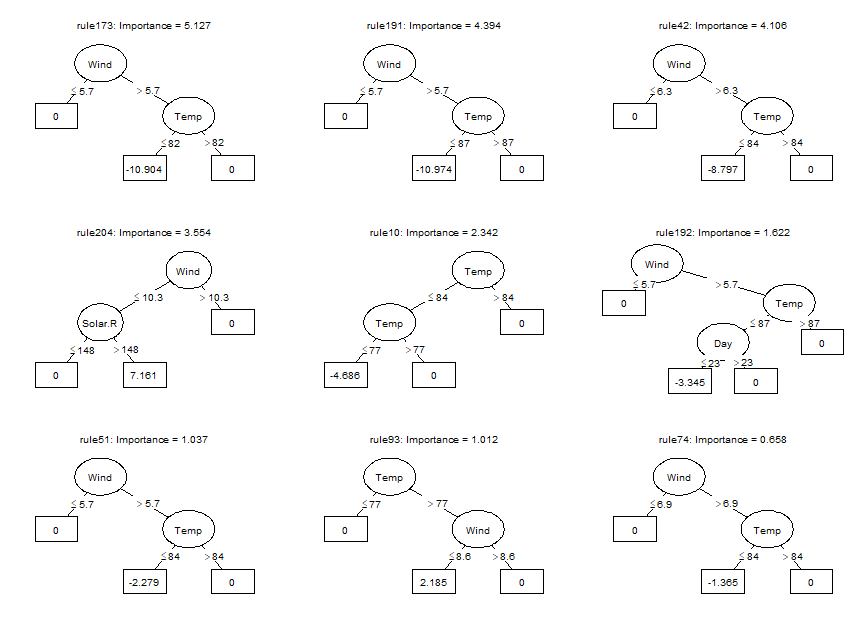
Using the plot method, we can plot the rules in the ensemble as simple
decision trees. Here, we will request the nine most important
baselearners through specification of the nterms argument. Through the
cex argument, we specify the size of the node and path labels:
plot(airq.ens, nterms = 9, cex = .5)Using the coef method, we can obtain the estimated coefficients for
each of the baselearners (we only print the first six terms here for
space considerations):
coefs <- coef(airq.ens)
coefs[1:6, ]
#> rule coefficient description
#> 201 (Intercept) 68.482704 1
#> 167 rule191 -10.973682 Wind > 5.7 & Temp <= 87
#> 150 rule173 -10.903855 Wind > 5.7 & Temp <= 82
#> 39 rule42 -8.797155 Wind > 6.3 & Temp <= 84
#> 179 rule204 7.161148 Wind <= 10.3 & Solar.R > 148
#> 10 rule10 -4.686461 Temp <= 84 & Temp <= 77We can generate predictions for new observations using the predict
method (only the first six predicted values are printed here for space
considerations):
predict(airq.ens, newdata = airq[1:6, ])
#> 1 2 3 4 7 8
#> 32.53896 24.22456 24.22456 24.22456 31.38570 24.22456Using function cvpre(), we can assess the expected prediction error of
the fitted PRE through k-fold cross validation (k = 10, by default,
which can be overridden through specification of the k argument):
set.seed(43)
airq.cv <- cvpre(airq.ens)
#> $MSE
#> MSE se
#> 369.92747 88.65343
#>
#> $MAE
#> MAE se
#> 13.666860 1.290329The results provide the mean squared error (MSE) and mean absolute error
(MAE) with their respective standard errors. These results are saved for
later use in aiq.cv$accuracy. The cross-validated predictions, which
can be used to compute alternative estimates of predictive accuracy, are
saved in airq.cv$cvpreds. The folds to which observations were
assigned are saved in airq.cv$fold_indicators.
For tuning the parameters of function pre() so as to obtain optimal
predictive accuracy, users are advised to use R package
caret. A tutorial is provided as a vignette, accessible by typing
vignette("tuning", package = "pre") in R or by going to
https://cran.r-project.org/package=pre/vignettes/missingness.html in a
browser and clicking on the corresponding link to the vignette.
Package pre provides several additional tools for interpretation
of the final ensemble. These may be especially helpful for complex
ensembles containing many rules and linear terms.
We can assess the relative importance of input variables as well as
baselearners using the importance() function:
imps <- importance(airq.ens, round = 4)As we already observed in the printed ensemble, the plotted variable
importances indicate that Temperature and Wind are most strongly
associated with Ozone levels. Solar.R and Day are also associated with
Ozone levels, but much less strongly. Variable Month is not plotted,
which means it obtained an importance of zero, indicating that it is not
associated with Ozone levels. We already observed this in the printed
ensemble: Month did not appear in any of the selected terms. The
variable and baselearner importances are saved for later use in
imps$varimps and imps$baseimps, respectively.
We can obtain explanations of the predictions for individual
observations using function explain():
par(mfrow = c(1, 2))
expl <- explain(airq.ens, newdata = airq[1:2, ], cex = .8)The values of the rules and linear terms for each observation are saved
in expl$predictors, their contributions in expl$contribution and the
predicted values in expl$predicted.value.
We can obtain partial dependence plots to assess the effect of single
predictor variables on the outcome using the singleplot() function:
singleplot(airq.ens, varname = "Temp")We can obtain partial dependence plots to assess the effects of pairs of
predictor variables on the outcome using the pairplot() function:
pairplot(airq.ens, varnames = c("Temp", "Wind"))Note that creating partial dependence plots is computationally intensive
and computation time will increase fast with increasing numbers of
observations and numbers of variables. Milborrow’s (2018) plotmo
package provides more efficient functions for plotting partial
dependence, which also support pre models.
If the final ensemble does not contain many terms, inspecting individual
rules and linear terms through the print method may be more
informative than partial dependence plots. One of the main advantages of
prediction rule ensembles is their interpretability: the predictive
model contains only simple functions of the predictor variables (rules
and linear terms), which are easy to grasp. Partial dependence plots are
often much more useful for interpretation of complex models, like random
forests for example.
We can assess the presence of interactions between the input variables
using the interact() and bsnullinteract() funtions. Function
bsnullinteract() computes null-interaction models (10, by default)
based on bootstrap-sampled and permuted datasets. Function interact()
computes interaction test statistics for each predictor variables
appearing in the specified ensemble. If null-interaction models are
provided through the nullmods argument, interaction test statistics
will also be computed for the null-interaction model, providing a
reference null distribution.
Note that computing null interaction models and interaction test statistics is computationally very intensive, so running the following code will take some time:
set.seed(44)
nullmods <- bsnullinteract(airq.ens)
int <- interact(airq.ens, nullmods = nullmods)The plotted variable interaction strengths indicate that Temperature and
Wind may be involved in interactions, as their observed interaction
strengths (darker grey) exceed the upper limit of the 90% confidence
interval (CI) of interaction stengths in the null interaction models
(lighter grey bar represents the median, error bars represent the 90%
CIs). The plot indicates that Solar.R and Day are not involved in any
interactions. Note that computation of null interaction models is
computationally intensive. A more reliable result can be obtained by
computing a larger number of boostrapped null interaction datasets, by
setting the nsamp argument of function bsnullinteract() to a larger
value (e.g., 100).
We can assess correlations between the baselearners appearing in the
ensemble using the corplot() function:
corplot(airq.ens)To obtain an optimal set of model-fitting parameters, package
caret Kuhn (2008) provides a method "pre". For a manual on how
to optimize the parameters of function pre using caret’s train
function, see the vignette on tuning:
vignette("tuning", package = "pre")or go to https://cran.r-project.org/package=pre/vignettes/tuning.html in a browser.
Some suggestions on how to deal with missing values are provided in the following vignette:
vignette("missingness", package = "pre")or go to https://cran.r-project.org/package=pre/vignettes/missingness.html in a browser.
When sparsity (i.e., a final ensemble comprising only few terms) is of central importance, the so-called relaxed lasso can be employed. It allows for retaining a pre-specified (low) number of terms, with adequate predictive accuracy. An introduction and tutorial is provided in the following vignette:
vignette("relaxed", package = "pre")An even more flexible ensembling approach is implemented in function
gpe(), which allows for fitting Generalized Prediction Ensembles: It
combines the MARS (multivariate Adaptive Splines) approach of Friedman
(1991) with the RuleFit approach of Friedman & Popescu (2008). In other
words, gpe() fits an ensemble composed of hinge functions (possibly
multivariate), prediction rules and linear functions of the predictor
variables. See the following example:
set.seed(42)
airq.gpe <- gpe(Ozone ~ ., data = airquality[complete.cases(airquality),],
base_learners = list(gpe_trees(), gpe_linear(), gpe_earth()))
airq.gpe
#>
#> Final ensemble with cv error within 1se of minimum:
#>
#> lambda = 3.229132
#> number of terms = 11
#> mean cv error (se) = 359.2623 (110.8863)
#>
#> cv error type : Mean-squared Error
#>
#> description coefficient
#> (Intercept) 65.52169488
#> Temp <= 77 -6.20973855
#> Wind <= 10.3 & Solar.R > 148 5.46410966
#> Wind > 5.7 & Temp <= 82 -8.06127415
#> Wind > 5.7 & Temp <= 84 -7.16921733
#> Wind > 5.7 & Temp <= 87 -8.04255471
#> Wind > 5.7 & Temp <= 87 & Day <= 23 -3.40525575
#> Wind > 6.3 & Temp <= 82 -2.71925536
#> Wind > 6.3 & Temp <= 84 -5.99085126
#> Wind > 6.9 & Temp <= 82 -0.04406376
#> Wind > 6.9 & Temp <= 84 -0.55827336
#> eTerm(Solar.R * h(9.7 - Wind), scale = 410) 9.91783320
#>
#> 'h' in the 'eTerm' indicates the hinge functionThe results indicate that several rules, a single hinge (linear spline) function, and no linear terms were selected for the final ensemble. The hinge function and its coefficient indicate that Ozone levels increase with increasing solar radiation and decreasing wind speeds. The prediction rules in the ensemble indicate a similar pattern.
I am very grateful to package co-author Benjamin Chistoffersen:
https://github.com/boennecd, who developed gpe and contributed
tremendously by improving functions, code and computational aspects.
Furthermore, I am grateful for the many helpful suggestions of Stephen
Milborrow, and for the contributions of Karl Holub
(https://github.com/holub008) and Advik Shreekumar
(https://github.com/adviksh).
Breiman, L., Friedman, J., Olshen, R., & Stone, C. (1984). Classification and regression trees. Boca Raton, FL: Chapman & Hall / CRC.
Fokkema, M. (2020). Fitting prediction rule ensembles with R package pre. Journal of Statistical Software, 92(12), 1–30. Retrieved from https://doi.org/10.18637/jss.v092.i12
Fokkema, M., & Strobl, C. (2020). Fitting prediction rule ensembles to psychological research data: An introduction and tutorial. Psychological Methods, 25(5), 636–652. https://doi.org/10.1037/met0000256
Friedman, J. (1991). Multivariate adaptive regression splines. The Annals of Statistics, 19, 1–67.
Friedman, J., & Popescu, B. (2008). Predictive learning via rule ensembles. The Annals of Applied Statistics, 2(3), 916–954.
Hothorn, T., Hornik, K., & Zeileis, A. (2006). Unbiased recursive partitioning: A conditional inference framework. Journal of Computational and Graphical Statistics, 15(3), 651–674.
Kuhn, M. (2008). Building predictive models in R using the caret package. Journal of Statistical Software, 28(5), 1–26.
Milborrow, S. (2018). plotmo: Plot a model’s residuals, response, and partial dependence plots. Retrieved from https://CRAN.R-project.org/package=plotmo
Zeileis, A., Hothorn, T., & Hornik, K. (2008). Model-based recursive partitioning. Journal of Computational and Graphical Statistics, 17(2), 492–514.