Tools for global optimization, attractor finding and energy landscape exploration.
Source code: https://github.com/pele-python/pele
Documentation: http://pele-python.github.io/pele/
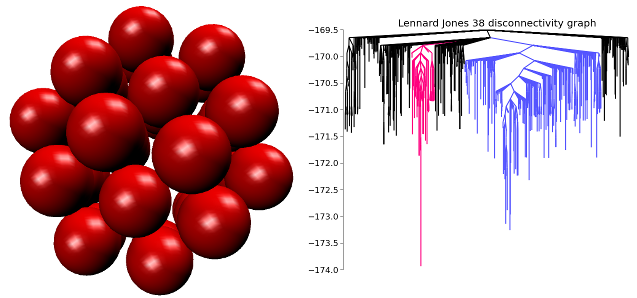
Images: The global minimum energy structure of a 38 atom Lennard-Jones cluster. On the right is a disconnectivity graph showing a visualization of the energy landscape. The competing low energy basins are shown in color.
pele started as a python partial-rewriting of GMIN, OPTIM, and PATHSAMPLE: fortran programs written by David Wales of Cambridge University and collaborators (http://www-wales.ch.cam.ac.uk/software.html).
The current version is being developed by the Martiniani group at New York University.
pele has tools for energy minimization, global optimization, saddle point (transition state) search, data analysis, visualization and much more. Some of the algorithms implemented are:
- Basinhopping global optimization
- Potentials (Lennard-Jones, Morse, Hertzian, etc.)
- LBFGS minimization (plus other minimizers)
- Attractor identification (Mixed Descent, CVODE)
- Single ended saddle point search: - Hybrid Eigenvector Following - Dimer method
- Double ended saddle point search - Nudged Elastic Band (NEB) - Doubly Nudged Elastic Band (DNEB)
- Disconnectivity Graph visualization
- Structure alignment algorithms
- Thermodynamics (e.g. heat capacity) via the Harmonic Superposition Approximation
- Transition rates analysis
- fortran compiler
- c compiler (gcc preferably)
- c++ compiler (g++ preferably)
- CMake (version 3.5 or higher)
On Ubuntu, the necessary software for compilation can be installed with sudo apt-get install gfortran gcc g++ cmake.
On older versions of Ubuntu you may need to provide a version number, and set the version used:
$ sudo apt install -y gcc-10 g++-10 gfortran-10 cmake # any gcc>5 $ sudo update-alternatives --install /usr/bin/gcc gcc /usr/bin/gcc-10 100 $ sudo update-alternatives --install /usr/bin/g++ g++ /usr/bin/g++-10 100 $ sudo update-alternatives --install /usr/bin/gfortran gfortran /usr/bin/gfortran-10 100
On Macs (for both Intel and Apple silicon), we recommend using homebrew to install the necessary software and libraries for compilation. Once homebrew is installed, use:
$ brew install gcc@13 cmake openblas gettext
Among other things, this will install version 13 of gcc and give you access to the gcc-13 and g++-13 commands. Be aware that Apple provides its own compilers under the commands gcc and g++ which, however, just run clang and not the GNU compilers. Since we do not support using the clang compilers at the moment, we have to make sure that the compilers installed by homebrew are used in the following. If you installed a different version of gcc, make sure to replace the gcc-13 and g++-13 parts accordingly.
- Eigen (http://eigen.tuxfamily.org)
- SUNDIALS (https://computing.llnl.gov/projects/sundials)
SUNDIALS and Eigen are automatically downloaded with git submodule update --init --recursive (which will also download GoogleTest for C++ tests)
an install script sun_inst.sh is provided for sundials in the install folder. Eigen can be installed by running the command cp -r eigen/Eigen install/include/ in the extern folder.
Run:
$ git submodule update --init --recursive $ cd extern $ ./sun_inst.sh release $ cp -r eigen/Eigen install/include/ $ cd ..
Use the commands for Ubuntu, however, set the correct compilers when running the install script sun_inst.sh by setting the CC and CXX environment variables. Also, make sure to use your current MacOs version as the deployment target:
$ MACOSX_DEPLOYMENT_TARGET=14.3 CC=gcc-13 CXX=g++-13 ./sun_inst.sh release
pele requires python 3.9 and the following packages
- numpy:
- We use numpy everywhere for doing numerical work. It also installs f2py which is used to compile fortran code into modules callable by python.
- scipy:
- For some of the optimizers and various scientific tools
- networkx:
- For graph functionality. https://networkx.lanl.gov
- cython (version 0.29.36):
- For calling C++ code from python for speed
- pyyaml:
- For reading and writing yaml files
- future:
- Used for upgrading from python 2 to python 3
- omp-thread-count:
- used to set the number of threads used by openmp
- matplotlib:
- For making plots (e.g. disconnectivity graphs)
- SQLAlchemy (version 1.4.51):
- For managing database of stationary points. http://www.sqlalchemy.org/
- munkres:
- For permutational alignment
- pyro4:
- For parallel jobs
- scikits.sparse: optional
- For use of sparse Cholesky decomposition methods when calculating rates
- pymol: optional
- For viewing molecular structures
- pytest: optional
- For running tests
We recommend installing all the above packages in a conda environment.
If you want to use the gui you will additionally need:
- qt4 and qt4 python bindings
- opengl python bindings
The Ubuntu packages (apt-get) for these are: python-qt4, python-opengl, and python-qt4-gl
In fedora Fedora (yum) you will want the packages: PyQt4, and PyOpenGl
We recommend to install Anaconda. On Ubuntu, set up a new conda environment using:
$ conda create -n myenv python=3.9 $ conda activate myenv $ conda install numpy scipy networkx matplotlib cython=0.29.36 $ conda install -c conda-forge sqlalchemy=1.4.51 munkres pyro4 scikit-sparse $ conda install -c conda-forge -c schrodinger pymol-bundle $ pip install pyyaml $ pip install omp-thread-count # for multi-threading $ pip install future # used for upgrading to python 3 $ pip install pytest # in case you want to ensure library runs correctly (optional)
On MacOs, follow the same commands but make sure that the installation of omp-thread-count uses the correct compiler by setting the CC environment variable:
$ CC=gcc-13 pip install omp-thread-count # for multi-threading
Also, note that the pymol-bundle package is not available on Apple silicon.
Compilation is required as many of the computationally intensive parts (especially potentials) are written in fortran and c++. Theoretically you should be able to use any compilers, but we mostly use gfortran and GCC, so it's the least likely to have problems. This package uses the standard python setup utility (distutils). The current installation procedure on Ubuntu is:
$ python setup_with_cmake.py develop
On MacOs, one has to set the deployment target according to the MacOs version again (the CC and CXX environment variables are set by the Python script):
$ MACOSX_DEPLOYMENT_TARGET=14.3 python3 setup_with_cmake.py develop
This compiles the extension modules and ensures that the python interpreter can find pele. You can also just compile the extension modules by using the command (possibly including the deployment target, if on MacOs):
$ python setup_with_cmake.py build_ext -i
Afterwards, make sure to add the install directory to your PYTHONPATH environment variable. To test whether your installation has worked correctly, run:
$ OMP_NUM_THREADS=1 pytest pele/
from the base directory. In order to install pele without attractor
identification support (i.e., without CVODE) use the
--with-cvode command-line option. For example, run:
$ python setup_with_cmake.py build_ext -i --with-cvode 0
Note that this will make some of the tests fail.
To check whether the code you're interested in works correctly you can run pytest
in the module you're interested in, for example, to check whether pele/utils is working correctly, run pytest pele/utils.
If building fails, run the following command to remove cached files before building again:
$ rm -rf build cythonize.dat CMakeCache.txt cmake_install.cmake
The C++ tests use GoogleTest. To run the tests, after running git submodule update --init --recursive to get the GoogleTest submodule if you haven't already, run:
$ cd cpp_tests/source $ cmake -DCMAKE_BUILD_TYPE=Debug . $ make -j8 $ ./test_main
On MacOs, use the same commands but make sure that cmake finds the correct GNU compilers and the OpenBLAS library:
$ cmake -DCMAKE_BUILD_TYPE=Debug -DCMAKE_C_COMPILER=gcc-13 -DCMAKE_CXX_COMPILER=g++-13 -DCMAKE_PREFIX_PATH=$(brew --prefix openblas) .
The python tests have originally been written using nose. But we have transitioned to using pytests. To run the tests, run:
$ pytest pele/
from the base directory.