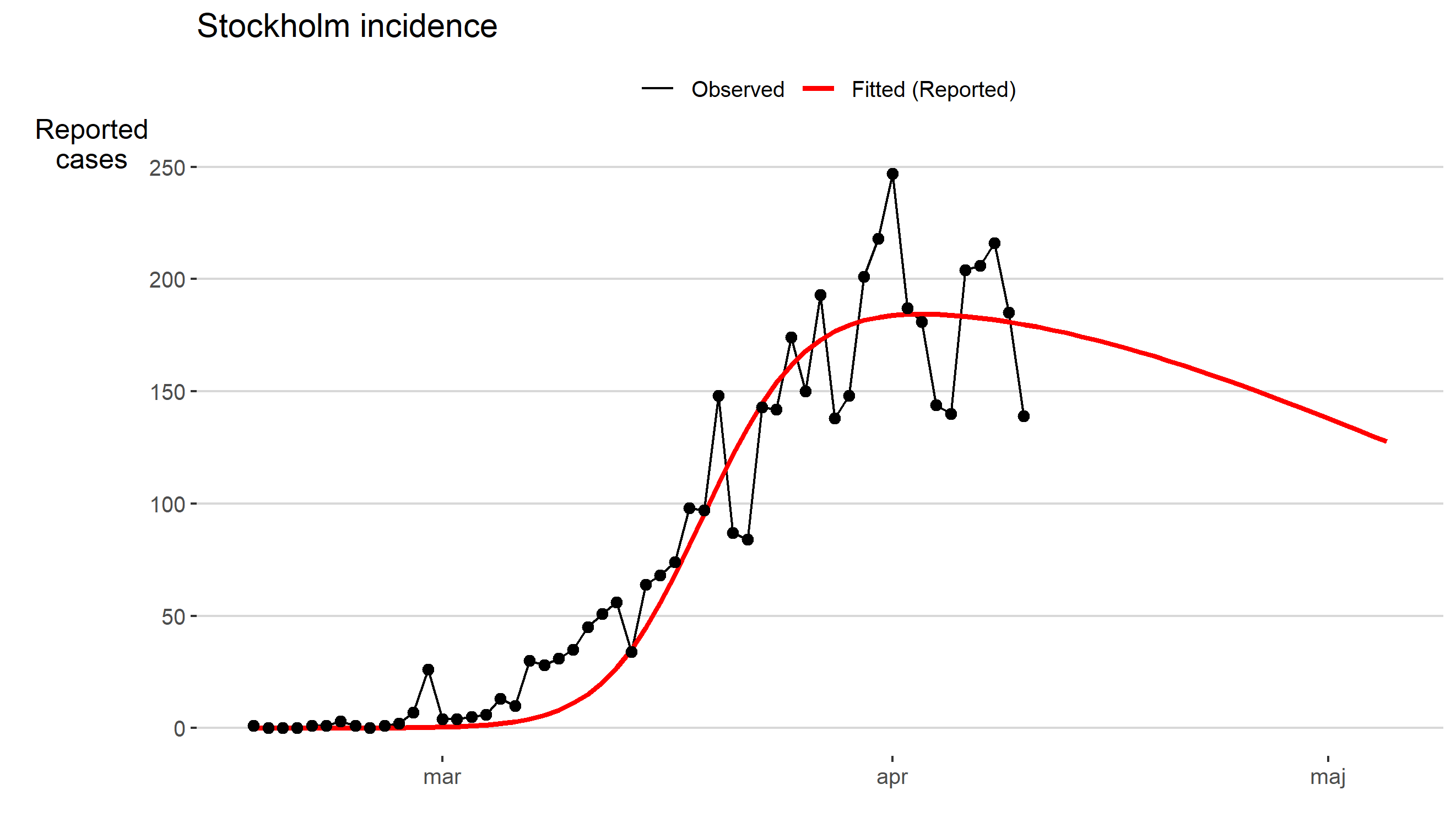
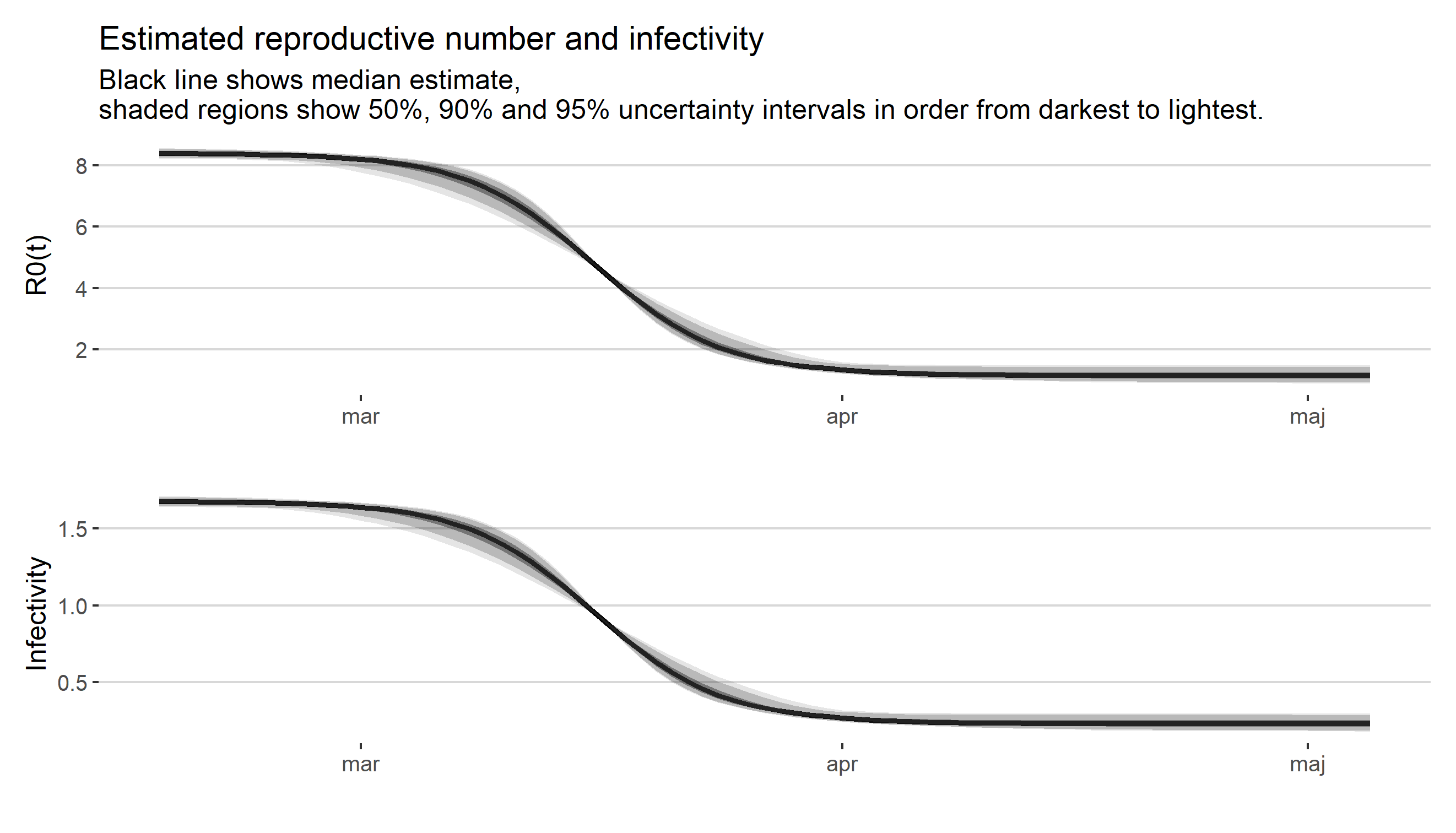
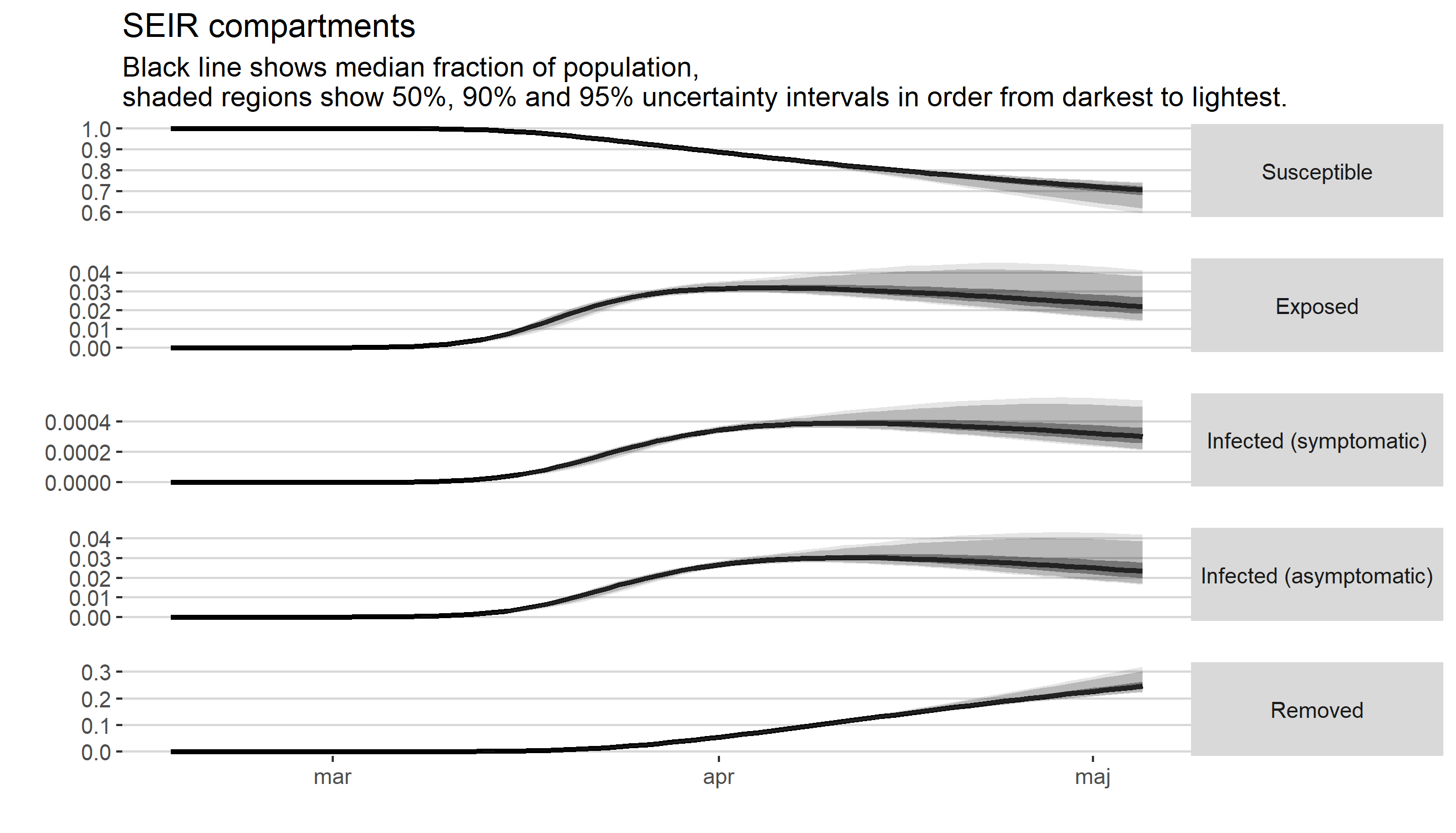
Skattning av peakdag och antal infekterade i covid-19-utbrottet i Stockholms län februari-april 2020.
This repo is a fork of Folkhälsomyndigheten’s repo containing the code for the SEIR model of the number of Covid-19 cases in Stockholm. I created this repo to understand their model better, find bugs or improvements to their code. I list my findings below.
- In the original code, parameters are drawn from a distribution to calculate uncertainty estimates. The implicit reason why one can do that is that the optimization procedure is a maximum likelihood estimation, and such estimators have an asymptotic normal distribution. However, this distribution is multivariate normal, but the original code draws each parmeter independently from each other. The code in this repo fixes this.
- Parameters are transformed to each lie on the real line before being
passed to
optim. This fixes the non-convergence that the original code sometimes displayed. - Code has been parallelized via the
furrrpackage where applicable, resulting in a pretty good speedup. - Uncertainty estimates are calculated for more quantities, such as the infectivity.
- Code now uses
tidyversepackages to a much greater extent. - Functions are documented (work in progress).
- Cleaner variable and function names.
- Nicer plots via
ggplot2.
Will explore model sensitivity to parameters gamma, eta and p_symp
here.
3622e30b12c46853c5fd0e4e42d2670975e0ae6e